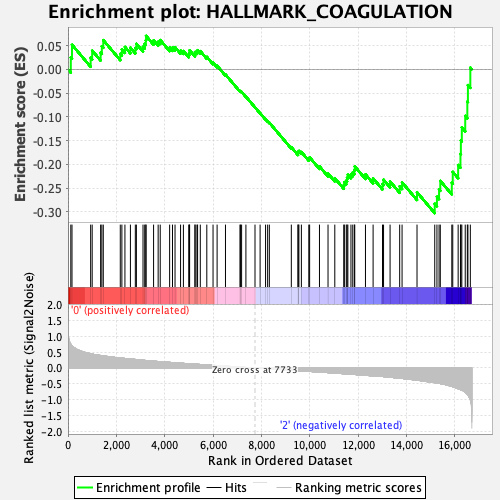
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.3029345 |
| Normalized Enrichment Score (NES) | -1.2117916 |
| Nominal p-value | 0.12104283 |
| FDR q-value | 0.66574866 |
| FWER p-Value | 0.922 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gsn | 116 | 0.751 | 0.0250 | No |
| 2 | P2ry1 | 165 | 0.696 | 0.0518 | No |
| 3 | Pecam1 | 937 | 0.450 | 0.0245 | No |
| 4 | Usp11 | 1000 | 0.438 | 0.0395 | No |
| 5 | Wdr1 | 1349 | 0.393 | 0.0353 | No |
| 6 | Plek | 1400 | 0.388 | 0.0488 | No |
| 7 | Trf | 1459 | 0.381 | 0.0616 | No |
| 8 | F10 | 2162 | 0.315 | 0.0327 | No |
| 9 | Olr1 | 2228 | 0.310 | 0.0420 | No |
| 10 | Ctsh | 2354 | 0.300 | 0.0473 | No |
| 11 | Itga2 | 2581 | 0.284 | 0.0457 | No |
| 12 | Itgb3 | 2784 | 0.269 | 0.0451 | No |
| 13 | Klf7 | 2828 | 0.266 | 0.0538 | No |
| 14 | Cd9 | 3102 | 0.243 | 0.0477 | No |
| 15 | Fyn | 3172 | 0.239 | 0.0538 | No |
| 16 | S100a13 | 3218 | 0.235 | 0.0611 | No |
| 17 | Pf4 | 3228 | 0.235 | 0.0705 | No |
| 18 | Comp | 3539 | 0.214 | 0.0610 | No |
| 19 | Ctsl | 3728 | 0.203 | 0.0583 | No |
| 20 | C3 | 3814 | 0.199 | 0.0617 | No |
| 21 | Ctsk | 4203 | 0.176 | 0.0459 | No |
| 22 | Proz | 4321 | 0.169 | 0.0460 | No |
| 23 | Gda | 4426 | 0.163 | 0.0467 | No |
| 24 | Gp1ba | 4655 | 0.151 | 0.0394 | No |
| 25 | Rac1 | 4772 | 0.145 | 0.0386 | No |
| 26 | Lamp2 | 4995 | 0.133 | 0.0309 | No |
| 27 | Masp2 | 5016 | 0.131 | 0.0353 | No |
| 28 | Thbs1 | 5029 | 0.131 | 0.0401 | No |
| 29 | Capn5 | 5250 | 0.118 | 0.0319 | No |
| 30 | Gng12 | 5255 | 0.118 | 0.0367 | No |
| 31 | Lgmn | 5332 | 0.115 | 0.0370 | No |
| 32 | Cfb | 5359 | 0.115 | 0.0403 | No |
| 33 | Gp9 | 5471 | 0.110 | 0.0383 | No |
| 34 | Plg | 5740 | 0.096 | 0.0263 | No |
| 35 | Anxa1 | 5998 | 0.083 | 0.0144 | No |
| 36 | Capn2 | 6166 | 0.075 | 0.0075 | No |
| 37 | Mmp8 | 6512 | 0.057 | -0.0109 | No |
| 38 | Fbn1 | 7113 | 0.027 | -0.0458 | No |
| 39 | Lta4h | 7130 | 0.026 | -0.0457 | No |
| 40 | C2 | 7179 | 0.024 | -0.0475 | No |
| 41 | Dusp6 | 7357 | 0.015 | -0.0575 | No |
| 42 | Mmp11 | 7733 | 0.000 | -0.0801 | No |
| 43 | Hpn | 7944 | -0.008 | -0.0924 | No |
| 44 | Lrp1 | 8170 | -0.019 | -0.1051 | No |
| 45 | C8g | 8256 | -0.023 | -0.1093 | No |
| 46 | Fn1 | 8326 | -0.027 | -0.1123 | No |
| 47 | Gnb2 | 9235 | -0.070 | -0.1640 | No |
| 48 | F2rl2 | 9512 | -0.084 | -0.1770 | No |
| 49 | C1qa | 9516 | -0.084 | -0.1736 | No |
| 50 | Csrp1 | 9548 | -0.085 | -0.1718 | No |
| 51 | Bmp1 | 9651 | -0.090 | -0.1741 | No |
| 52 | Serpinc1 | 9958 | -0.106 | -0.1880 | No |
| 53 | Dpp4 | 9992 | -0.107 | -0.1854 | No |
| 54 | Mmp2 | 10399 | -0.128 | -0.2044 | No |
| 55 | Klk8 | 10753 | -0.146 | -0.2195 | No |
| 56 | Msrb2 | 11033 | -0.163 | -0.2293 | No |
| 57 | Thbd | 11404 | -0.181 | -0.2439 | No |
| 58 | Pef1 | 11428 | -0.182 | -0.2375 | No |
| 59 | Sh2b2 | 11507 | -0.186 | -0.2343 | No |
| 60 | Arf4 | 11541 | -0.189 | -0.2282 | No |
| 61 | Hmgcs2 | 11573 | -0.190 | -0.2220 | No |
| 62 | Mmp14 | 11701 | -0.198 | -0.2212 | No |
| 63 | Adam9 | 11776 | -0.201 | -0.2171 | No |
| 64 | Ctsb | 11842 | -0.205 | -0.2123 | No |
| 65 | Rabif | 11858 | -0.205 | -0.2045 | No |
| 66 | Casp9 | 12301 | -0.231 | -0.2212 | No |
| 67 | Sirt2 | 12617 | -0.246 | -0.2297 | No |
| 68 | Pdgfb | 13006 | -0.269 | -0.2416 | No |
| 69 | Mmp15 | 13049 | -0.271 | -0.2326 | No |
| 70 | Prep | 13319 | -0.292 | -0.2363 | No |
| 71 | Ctso | 13716 | -0.323 | -0.2464 | No |
| 72 | Serpinb2 | 13815 | -0.330 | -0.2382 | No |
| 73 | Cpq | 14433 | -0.386 | -0.2589 | No |
| 74 | Vwf | 15166 | -0.463 | -0.2832 | Yes |
| 75 | Cpn1 | 15257 | -0.475 | -0.2684 | Yes |
| 76 | Furin | 15345 | -0.485 | -0.2530 | Yes |
| 77 | S100a1 | 15396 | -0.492 | -0.2350 | Yes |
| 78 | Apoc1 | 15872 | -0.580 | -0.2389 | Yes |
| 79 | F8 | 15911 | -0.590 | -0.2160 | Yes |
| 80 | Pros1 | 16134 | -0.653 | -0.2015 | Yes |
| 81 | Iscu | 16230 | -0.681 | -0.1782 | Yes |
| 82 | Maff | 16247 | -0.689 | -0.1498 | Yes |
| 83 | Ctse | 16286 | -0.702 | -0.1222 | Yes |
| 84 | Ang | 16427 | -0.764 | -0.0981 | Yes |
| 85 | Rapgef3 | 16514 | -0.829 | -0.0679 | Yes |
| 86 | Timp3 | 16538 | -0.847 | -0.0332 | Yes |
| 87 | Hnf4a | 16638 | -1.005 | 0.0037 | Yes |

