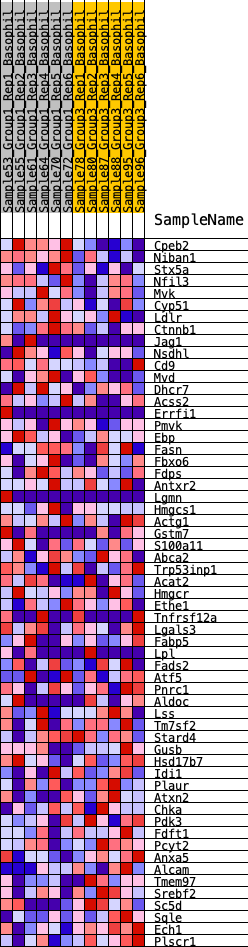
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
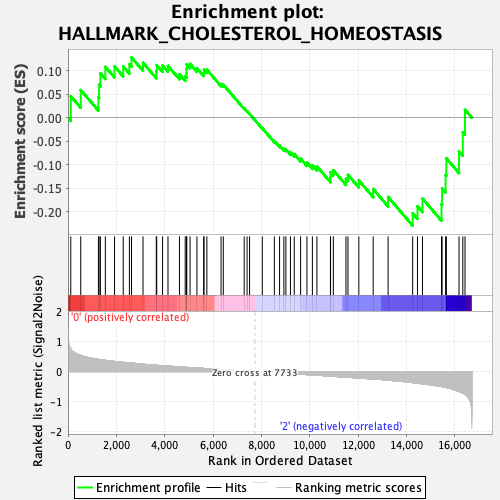
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.22985168 |
| Normalized Enrichment Score (NES) | -0.87733334 |
| Nominal p-value | 0.75232774 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpeb2 | 115 | 0.752 | 0.0459 | No |
| 2 | Niban1 | 527 | 0.538 | 0.0590 | No |
| 3 | Stx5a | 1258 | 0.404 | 0.0435 | No |
| 4 | Nfil3 | 1285 | 0.401 | 0.0701 | No |
| 5 | Mvk | 1340 | 0.395 | 0.0946 | No |
| 6 | Cyp51 | 1544 | 0.374 | 0.1086 | No |
| 7 | Ldlr | 1925 | 0.335 | 0.1093 | No |
| 8 | Ctnnb1 | 2284 | 0.306 | 0.1092 | No |
| 9 | Jag1 | 2539 | 0.285 | 0.1140 | No |
| 10 | Nsdhl | 2627 | 0.280 | 0.1285 | No |
| 11 | Cd9 | 3102 | 0.243 | 0.1171 | No |
| 12 | Mvd | 3650 | 0.207 | 0.0987 | No |
| 13 | Dhcr7 | 3669 | 0.206 | 0.1121 | No |
| 14 | Acss2 | 3910 | 0.193 | 0.1113 | No |
| 15 | Errfi1 | 4135 | 0.179 | 0.1104 | No |
| 16 | Pmvk | 4609 | 0.153 | 0.0927 | No |
| 17 | Ebp | 4847 | 0.141 | 0.0884 | No |
| 18 | Fasn | 4901 | 0.138 | 0.0948 | No |
| 19 | Fbxo6 | 4904 | 0.138 | 0.1044 | No |
| 20 | Fdps | 4909 | 0.137 | 0.1138 | No |
| 21 | Antxr2 | 5047 | 0.130 | 0.1147 | No |
| 22 | Lgmn | 5332 | 0.115 | 0.1057 | No |
| 23 | Hmgcs1 | 5611 | 0.103 | 0.0962 | No |
| 24 | Actg1 | 5624 | 0.102 | 0.1026 | No |
| 25 | Gstm7 | 5742 | 0.096 | 0.1023 | No |
| 26 | S100a11 | 6329 | 0.066 | 0.0717 | No |
| 27 | Abca2 | 6422 | 0.061 | 0.0705 | No |
| 28 | Trp53inp1 | 7286 | 0.019 | 0.0200 | No |
| 29 | Acat2 | 7405 | 0.013 | 0.0138 | No |
| 30 | Hmgcr | 7509 | 0.009 | 0.0082 | No |
| 31 | Ethe1 | 8035 | -0.013 | -0.0224 | No |
| 32 | Tnfrsf12a | 8533 | -0.036 | -0.0498 | No |
| 33 | Lgals3 | 8749 | -0.046 | -0.0595 | No |
| 34 | Fabp5 | 8927 | -0.054 | -0.0663 | No |
| 35 | Lpl | 9011 | -0.059 | -0.0671 | No |
| 36 | Fads2 | 9199 | -0.068 | -0.0736 | No |
| 37 | Atf5 | 9356 | -0.076 | -0.0776 | No |
| 38 | Pnrc1 | 9619 | -0.088 | -0.0872 | No |
| 39 | Aldoc | 9880 | -0.102 | -0.0957 | No |
| 40 | Lss | 10106 | -0.114 | -0.1012 | No |
| 41 | Tm7sf2 | 10294 | -0.123 | -0.1038 | No |
| 42 | Stard4 | 10853 | -0.151 | -0.1267 | No |
| 43 | Gusb | 10857 | -0.152 | -0.1162 | No |
| 44 | Hsd17b7 | 10972 | -0.158 | -0.1120 | No |
| 45 | Idi1 | 11495 | -0.186 | -0.1303 | No |
| 46 | Plaur | 11578 | -0.191 | -0.1218 | No |
| 47 | Atxn2 | 12027 | -0.215 | -0.1336 | No |
| 48 | Chka | 12621 | -0.247 | -0.1519 | No |
| 49 | Pdk3 | 13237 | -0.285 | -0.1689 | No |
| 50 | Fdft1 | 14253 | -0.369 | -0.2039 | Yes |
| 51 | Pcyt2 | 14451 | -0.386 | -0.1886 | Yes |
| 52 | Anxa5 | 14659 | -0.411 | -0.1722 | Yes |
| 53 | Alcam | 15447 | -0.499 | -0.1845 | Yes |
| 54 | Tmem97 | 15470 | -0.502 | -0.1505 | Yes |
| 55 | Srebf2 | 15615 | -0.527 | -0.1221 | Yes |
| 56 | Sc5d | 15646 | -0.532 | -0.0866 | Yes |
| 57 | Sqle | 16170 | -0.660 | -0.0716 | Yes |
| 58 | Ech1 | 16327 | -0.713 | -0.0310 | Yes |
| 59 | Plscr1 | 16418 | -0.758 | 0.0169 | Yes |