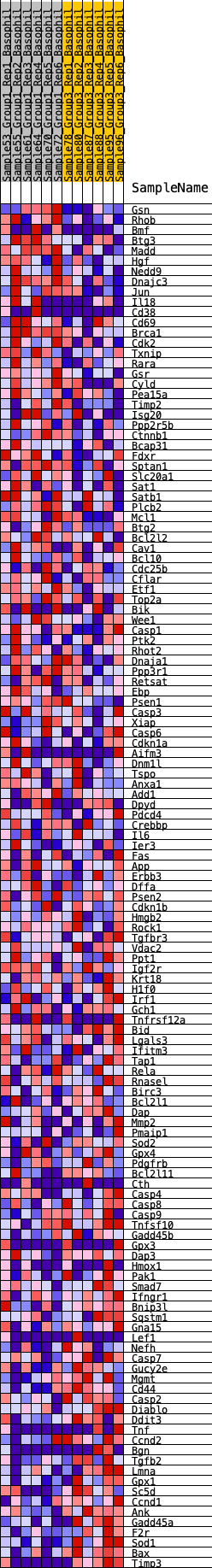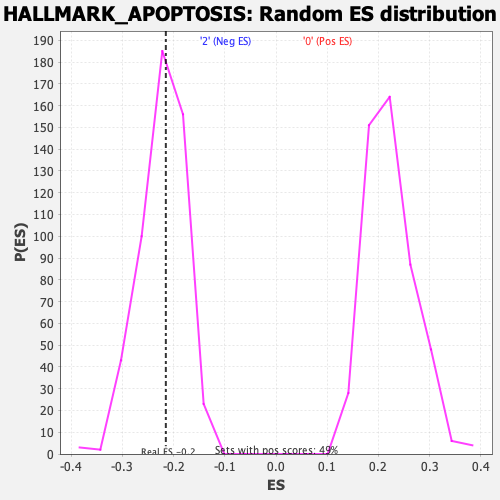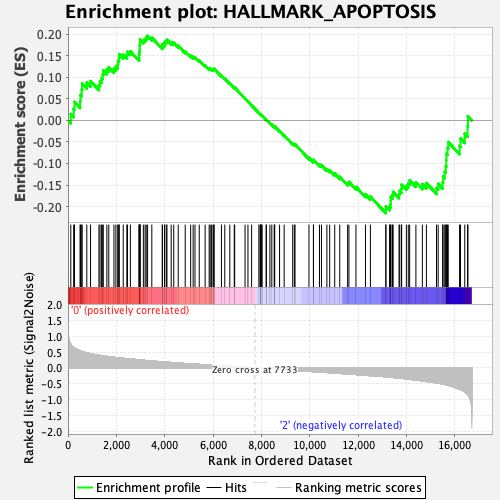
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.2151333 |
| Normalized Enrichment Score (NES) | -0.96880543 |
| Nominal p-value | 0.5234375 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gsn | 116 | 0.751 | 0.0146 | No |
| 2 | Rhob | 235 | 0.651 | 0.0262 | No |
| 3 | Bmf | 268 | 0.633 | 0.0426 | No |
| 4 | Btg3 | 500 | 0.546 | 0.0443 | No |
| 5 | Madd | 514 | 0.542 | 0.0592 | No |
| 6 | Hgf | 568 | 0.526 | 0.0711 | No |
| 7 | Nedd9 | 583 | 0.522 | 0.0853 | No |
| 8 | Dnajc3 | 777 | 0.475 | 0.0873 | No |
| 9 | Jun | 933 | 0.451 | 0.0910 | No |
| 10 | Il18 | 1280 | 0.402 | 0.0816 | No |
| 11 | Cd38 | 1323 | 0.396 | 0.0905 | No |
| 12 | Cd69 | 1388 | 0.388 | 0.0978 | No |
| 13 | Brca1 | 1436 | 0.384 | 0.1060 | No |
| 14 | Cdk2 | 1457 | 0.381 | 0.1158 | No |
| 15 | Txnip | 1607 | 0.366 | 0.1174 | No |
| 16 | Rara | 1687 | 0.357 | 0.1229 | No |
| 17 | Gsr | 1900 | 0.337 | 0.1198 | No |
| 18 | Cyld | 1981 | 0.330 | 0.1245 | No |
| 19 | Pea15a | 2061 | 0.324 | 0.1290 | No |
| 20 | Timp2 | 2069 | 0.324 | 0.1379 | No |
| 21 | Isg20 | 2108 | 0.320 | 0.1448 | No |
| 22 | Ppp2r5b | 2117 | 0.319 | 0.1535 | No |
| 23 | Ctnnb1 | 2284 | 0.306 | 0.1523 | No |
| 24 | Bcap31 | 2434 | 0.294 | 0.1518 | No |
| 25 | Fdxr | 2461 | 0.292 | 0.1586 | No |
| 26 | Sptan1 | 2579 | 0.284 | 0.1597 | No |
| 27 | Slc20a1 | 2939 | 0.255 | 0.1454 | No |
| 28 | Sat1 | 2941 | 0.255 | 0.1527 | No |
| 29 | Satb1 | 2953 | 0.254 | 0.1593 | No |
| 30 | Plcb2 | 2966 | 0.253 | 0.1659 | No |
| 31 | Mcl1 | 2967 | 0.253 | 0.1732 | No |
| 32 | Btg2 | 2976 | 0.253 | 0.1800 | No |
| 33 | Bcl2l2 | 2978 | 0.253 | 0.1872 | No |
| 34 | Cav1 | 3116 | 0.242 | 0.1859 | No |
| 35 | Bcl10 | 3187 | 0.238 | 0.1885 | No |
| 36 | Cdc25b | 3234 | 0.234 | 0.1925 | No |
| 37 | Cflar | 3291 | 0.231 | 0.1958 | No |
| 38 | Etf1 | 3466 | 0.219 | 0.1916 | No |
| 39 | Top2a | 3899 | 0.194 | 0.1711 | No |
| 40 | Bik | 3904 | 0.193 | 0.1764 | No |
| 41 | Wee1 | 3991 | 0.188 | 0.1766 | No |
| 42 | Casp1 | 3998 | 0.188 | 0.1817 | No |
| 43 | Ptk2 | 4065 | 0.184 | 0.1830 | No |
| 44 | Rhot2 | 4092 | 0.182 | 0.1867 | No |
| 45 | Dnaja1 | 4267 | 0.173 | 0.1812 | No |
| 46 | Ppp3r1 | 4373 | 0.166 | 0.1796 | No |
| 47 | Retsat | 4558 | 0.156 | 0.1730 | No |
| 48 | Ebp | 4847 | 0.141 | 0.1597 | No |
| 49 | Psen1 | 5072 | 0.128 | 0.1498 | No |
| 50 | Casp3 | 5175 | 0.123 | 0.1472 | No |
| 51 | Xiap | 5252 | 0.118 | 0.1460 | No |
| 52 | Casp6 | 5427 | 0.112 | 0.1387 | No |
| 53 | Cdkn1a | 5671 | 0.099 | 0.1269 | No |
| 54 | Aifm3 | 5838 | 0.091 | 0.1195 | No |
| 55 | Dnm1l | 5880 | 0.089 | 0.1196 | No |
| 56 | Tspo | 5938 | 0.087 | 0.1187 | No |
| 57 | Anxa1 | 5998 | 0.083 | 0.1175 | No |
| 58 | Add1 | 6022 | 0.082 | 0.1185 | No |
| 59 | Dpyd | 6046 | 0.081 | 0.1194 | No |
| 60 | Pdcd4 | 6345 | 0.066 | 0.1033 | No |
| 61 | Crebbp | 6483 | 0.058 | 0.0967 | No |
| 62 | Il6 | 6691 | 0.048 | 0.0856 | No |
| 63 | Ier3 | 6875 | 0.039 | 0.0757 | No |
| 64 | Fas | 6893 | 0.038 | 0.0758 | No |
| 65 | App | 7321 | 0.017 | 0.0505 | No |
| 66 | Erbb3 | 7442 | 0.011 | 0.0436 | No |
| 67 | Dffa | 7593 | 0.005 | 0.0347 | No |
| 68 | Psen2 | 7893 | -0.006 | 0.0168 | No |
| 69 | Cdkn1b | 7942 | -0.008 | 0.0141 | No |
| 70 | Hmgb2 | 7967 | -0.009 | 0.0129 | No |
| 71 | Rock1 | 7973 | -0.009 | 0.0129 | No |
| 72 | Tgfbr3 | 7976 | -0.010 | 0.0130 | No |
| 73 | Vdac2 | 7994 | -0.011 | 0.0123 | No |
| 74 | Ppt1 | 8029 | -0.012 | 0.0106 | No |
| 75 | Igf2r | 8198 | -0.020 | 0.0011 | No |
| 76 | Krt18 | 8199 | -0.020 | 0.0017 | No |
| 77 | H1f0 | 8350 | -0.028 | -0.0066 | No |
| 78 | Irf1 | 8428 | -0.031 | -0.0103 | No |
| 79 | Gch1 | 8532 | -0.036 | -0.0155 | No |
| 80 | Tnfrsf12a | 8533 | -0.036 | -0.0145 | No |
| 81 | Bid | 8548 | -0.037 | -0.0143 | No |
| 82 | Lgals3 | 8749 | -0.046 | -0.0250 | No |
| 83 | Ifitm3 | 8939 | -0.055 | -0.0349 | No |
| 84 | Tap1 | 9292 | -0.073 | -0.0540 | No |
| 85 | Rela | 9358 | -0.076 | -0.0557 | No |
| 86 | Rnasel | 9389 | -0.078 | -0.0553 | No |
| 87 | Birc3 | 9965 | -0.106 | -0.0870 | No |
| 88 | Bcl2l1 | 10149 | -0.116 | -0.0947 | No |
| 89 | Dap | 10151 | -0.116 | -0.0914 | No |
| 90 | Mmp2 | 10399 | -0.128 | -0.1026 | No |
| 91 | Pmaip1 | 10482 | -0.131 | -0.1038 | No |
| 92 | Sod2 | 10706 | -0.143 | -0.1131 | No |
| 93 | Gpx4 | 10821 | -0.150 | -0.1157 | No |
| 94 | Pdgfrb | 11030 | -0.162 | -0.1236 | No |
| 95 | Bcl2l11 | 11238 | -0.172 | -0.1311 | No |
| 96 | Cth | 11563 | -0.190 | -0.1452 | No |
| 97 | Casp4 | 11617 | -0.193 | -0.1429 | No |
| 98 | Casp8 | 11908 | -0.208 | -0.1544 | No |
| 99 | Casp9 | 12301 | -0.231 | -0.1714 | No |
| 100 | Tnfsf10 | 12505 | -0.241 | -0.1767 | No |
| 101 | Gadd45b | 13143 | -0.278 | -0.2071 | Yes |
| 102 | Gpx3 | 13146 | -0.279 | -0.1992 | Yes |
| 103 | Dap3 | 13296 | -0.290 | -0.1999 | Yes |
| 104 | Hmox1 | 13340 | -0.293 | -0.1940 | Yes |
| 105 | Pak1 | 13348 | -0.294 | -0.1860 | Yes |
| 106 | Smad7 | 13350 | -0.294 | -0.1776 | Yes |
| 107 | Ifngr1 | 13419 | -0.300 | -0.1730 | Yes |
| 108 | Bnip3l | 13445 | -0.301 | -0.1659 | Yes |
| 109 | Sqstm1 | 13689 | -0.321 | -0.1713 | Yes |
| 110 | Gna15 | 13719 | -0.324 | -0.1637 | Yes |
| 111 | Lef1 | 13790 | -0.329 | -0.1585 | Yes |
| 112 | Nefh | 13797 | -0.329 | -0.1494 | Yes |
| 113 | Casp7 | 13992 | -0.345 | -0.1512 | Yes |
| 114 | Gucy2e | 14072 | -0.351 | -0.1458 | Yes |
| 115 | Mgmt | 14129 | -0.356 | -0.1389 | Yes |
| 116 | Cd44 | 14387 | -0.382 | -0.1435 | Yes |
| 117 | Casp2 | 14654 | -0.410 | -0.1477 | Yes |
| 118 | Diablo | 14821 | -0.428 | -0.1454 | Yes |
| 119 | Ddit3 | 15239 | -0.472 | -0.1570 | Yes |
| 120 | Tnf | 15307 | -0.481 | -0.1472 | Yes |
| 121 | Ccnd2 | 15493 | -0.506 | -0.1438 | Yes |
| 122 | Bgn | 15511 | -0.508 | -0.1302 | Yes |
| 123 | Tgfb2 | 15580 | -0.521 | -0.1193 | Yes |
| 124 | Lmna | 15621 | -0.528 | -0.1065 | Yes |
| 125 | Gpx1 | 15644 | -0.532 | -0.0925 | Yes |
| 126 | Sc5d | 15646 | -0.532 | -0.0773 | Yes |
| 127 | Ccnd1 | 15695 | -0.543 | -0.0645 | Yes |
| 128 | Ank | 15723 | -0.548 | -0.0504 | Yes |
| 129 | Gadd45a | 16193 | -0.669 | -0.0594 | Yes |
| 130 | F2r | 16232 | -0.682 | -0.0421 | Yes |
| 131 | Sod1 | 16408 | -0.752 | -0.0310 | Yes |
| 132 | Bax | 16524 | -0.835 | -0.0139 | Yes |
| 133 | Timp3 | 16538 | -0.847 | 0.0097 | Yes |