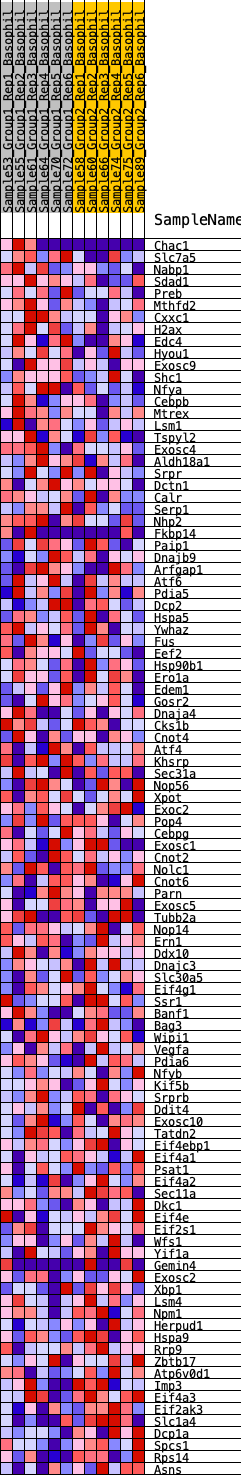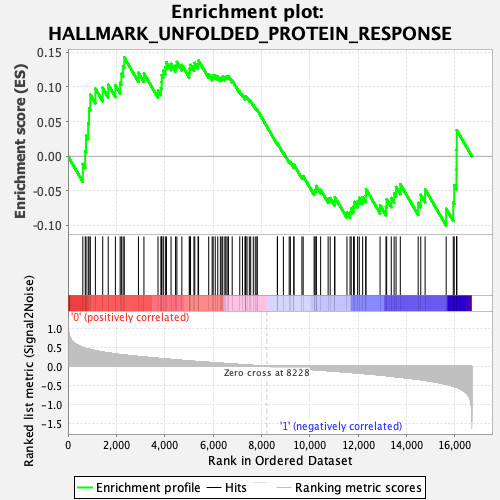
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.142456 |
| Normalized Enrichment Score (NES) | 0.6032987 |
| Nominal p-value | 0.9507187 |
| FDR q-value | 0.974808 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Chac1 | 613 | 0.500 | -0.0112 | Yes |
| 2 | Slc7a5 | 713 | 0.477 | 0.0073 | Yes |
| 3 | Nabp1 | 746 | 0.472 | 0.0296 | Yes |
| 4 | Sdad1 | 834 | 0.459 | 0.0480 | Yes |
| 5 | Preb | 870 | 0.452 | 0.0691 | Yes |
| 6 | Mthfd2 | 923 | 0.444 | 0.0888 | Yes |
| 7 | Cxxc1 | 1132 | 0.412 | 0.0974 | Yes |
| 8 | H2ax | 1436 | 0.377 | 0.0985 | Yes |
| 9 | Edc4 | 1663 | 0.353 | 0.1031 | Yes |
| 10 | Hyou1 | 1959 | 0.326 | 0.1021 | Yes |
| 11 | Exosc9 | 2157 | 0.309 | 0.1061 | Yes |
| 12 | Shc1 | 2207 | 0.306 | 0.1189 | Yes |
| 13 | Nfya | 2278 | 0.300 | 0.1301 | Yes |
| 14 | Cebpb | 2327 | 0.296 | 0.1425 | Yes |
| 15 | Mtrex | 2913 | 0.258 | 0.1205 | No |
| 16 | Lsm1 | 3140 | 0.244 | 0.1194 | No |
| 17 | Tspyl2 | 3722 | 0.209 | 0.0952 | No |
| 18 | Exosc4 | 3842 | 0.203 | 0.0984 | No |
| 19 | Aldh18a1 | 3870 | 0.201 | 0.1071 | No |
| 20 | Srpr | 3879 | 0.201 | 0.1170 | No |
| 21 | Dctn1 | 3941 | 0.198 | 0.1235 | No |
| 22 | Calr | 4025 | 0.193 | 0.1284 | No |
| 23 | Serp1 | 4071 | 0.190 | 0.1354 | No |
| 24 | Nhp2 | 4261 | 0.180 | 0.1333 | No |
| 25 | Fkbp14 | 4448 | 0.168 | 0.1307 | No |
| 26 | Paip1 | 4502 | 0.166 | 0.1361 | No |
| 27 | Dnajb9 | 4708 | 0.155 | 0.1317 | No |
| 28 | Arfgap1 | 5014 | 0.140 | 0.1205 | No |
| 29 | Atf6 | 5034 | 0.139 | 0.1265 | No |
| 30 | Pdia5 | 5067 | 0.138 | 0.1317 | No |
| 31 | Dcp2 | 5208 | 0.131 | 0.1300 | No |
| 32 | Hspa5 | 5243 | 0.129 | 0.1346 | No |
| 33 | Ywhaz | 5378 | 0.123 | 0.1329 | No |
| 34 | Fus | 5399 | 0.123 | 0.1380 | No |
| 35 | Eef2 | 5819 | 0.103 | 0.1180 | No |
| 36 | Hsp90b1 | 5962 | 0.097 | 0.1145 | No |
| 37 | Ero1a | 6003 | 0.095 | 0.1169 | No |
| 38 | Edem1 | 6089 | 0.091 | 0.1165 | No |
| 39 | Gosr2 | 6193 | 0.087 | 0.1148 | No |
| 40 | Dnaja4 | 6300 | 0.082 | 0.1126 | No |
| 41 | Cks1b | 6364 | 0.079 | 0.1128 | No |
| 42 | Cnot4 | 6400 | 0.078 | 0.1147 | No |
| 43 | Atf4 | 6487 | 0.074 | 0.1134 | No |
| 44 | Khsrp | 6528 | 0.073 | 0.1147 | No |
| 45 | Sec31a | 6580 | 0.071 | 0.1153 | No |
| 46 | Nop56 | 6635 | 0.068 | 0.1155 | No |
| 47 | Xpot | 6790 | 0.061 | 0.1094 | No |
| 48 | Exoc2 | 7103 | 0.046 | 0.0930 | No |
| 49 | Pop4 | 7214 | 0.042 | 0.0885 | No |
| 50 | Cebpg | 7323 | 0.038 | 0.0840 | No |
| 51 | Exosc1 | 7332 | 0.037 | 0.0854 | No |
| 52 | Cnot2 | 7355 | 0.037 | 0.0860 | No |
| 53 | Nolc1 | 7403 | 0.035 | 0.0849 | No |
| 54 | Cnot6 | 7498 | 0.031 | 0.0808 | No |
| 55 | Parn | 7555 | 0.028 | 0.0789 | No |
| 56 | Exosc5 | 7663 | 0.023 | 0.0737 | No |
| 57 | Tubb2a | 7743 | 0.020 | 0.0699 | No |
| 58 | Nop14 | 7816 | 0.017 | 0.0665 | No |
| 59 | Ern1 | 7822 | 0.017 | 0.0671 | No |
| 60 | Ddx10 | 8657 | -0.017 | 0.0177 | No |
| 61 | Dnajc3 | 8662 | -0.017 | 0.0183 | No |
| 62 | Slc30a5 | 8907 | -0.026 | 0.0050 | No |
| 63 | Eif4g1 | 9147 | -0.038 | -0.0075 | No |
| 64 | Ssr1 | 9195 | -0.040 | -0.0083 | No |
| 65 | Banf1 | 9340 | -0.047 | -0.0146 | No |
| 66 | Bag3 | 9343 | -0.047 | -0.0123 | No |
| 67 | Wipi1 | 9674 | -0.062 | -0.0290 | No |
| 68 | Vegfa | 9725 | -0.064 | -0.0287 | No |
| 69 | Pdia6 | 10178 | -0.086 | -0.0515 | No |
| 70 | Nfyb | 10213 | -0.088 | -0.0490 | No |
| 71 | Kif5b | 10262 | -0.090 | -0.0473 | No |
| 72 | Srprb | 10268 | -0.090 | -0.0430 | No |
| 73 | Ddit4 | 10447 | -0.099 | -0.0486 | No |
| 74 | Exosc10 | 10757 | -0.114 | -0.0614 | No |
| 75 | Tatdn2 | 10842 | -0.117 | -0.0604 | No |
| 76 | Eif4ebp1 | 11024 | -0.125 | -0.0649 | No |
| 77 | Eif4a1 | 11037 | -0.126 | -0.0592 | No |
| 78 | Psat1 | 11536 | -0.152 | -0.0814 | No |
| 79 | Eif4a2 | 11665 | -0.158 | -0.0809 | No |
| 80 | Sec11a | 11712 | -0.160 | -0.0755 | No |
| 81 | Dkc1 | 11808 | -0.166 | -0.0727 | No |
| 82 | Eif4e | 11840 | -0.168 | -0.0659 | No |
| 83 | Eif2s1 | 11977 | -0.173 | -0.0652 | No |
| 84 | Wfs1 | 12045 | -0.177 | -0.0601 | No |
| 85 | Yif1a | 12185 | -0.185 | -0.0590 | No |
| 86 | Gemin4 | 12313 | -0.192 | -0.0568 | No |
| 87 | Exosc2 | 12323 | -0.192 | -0.0475 | No |
| 88 | Xbp1 | 12905 | -0.226 | -0.0708 | No |
| 89 | Lsm4 | 13150 | -0.242 | -0.0731 | No |
| 90 | Npm1 | 13178 | -0.244 | -0.0622 | No |
| 91 | Herpud1 | 13368 | -0.258 | -0.0603 | No |
| 92 | Hspa9 | 13487 | -0.267 | -0.0537 | No |
| 93 | Rrp9 | 13566 | -0.273 | -0.0444 | No |
| 94 | Zbtb17 | 13742 | -0.284 | -0.0404 | No |
| 95 | Atp6v0d1 | 14481 | -0.341 | -0.0673 | No |
| 96 | Imp3 | 14588 | -0.350 | -0.0557 | No |
| 97 | Eif4a3 | 14768 | -0.365 | -0.0478 | No |
| 98 | Eif2ak3 | 15639 | -0.472 | -0.0759 | No |
| 99 | Slc1a4 | 15932 | -0.522 | -0.0667 | No |
| 100 | Dcp1a | 15972 | -0.529 | -0.0419 | No |
| 101 | Spcs1 | 16067 | -0.553 | -0.0191 | No |
| 102 | Rps14 | 16070 | -0.554 | 0.0093 | No |
| 103 | Asns | 16078 | -0.556 | 0.0374 | No |