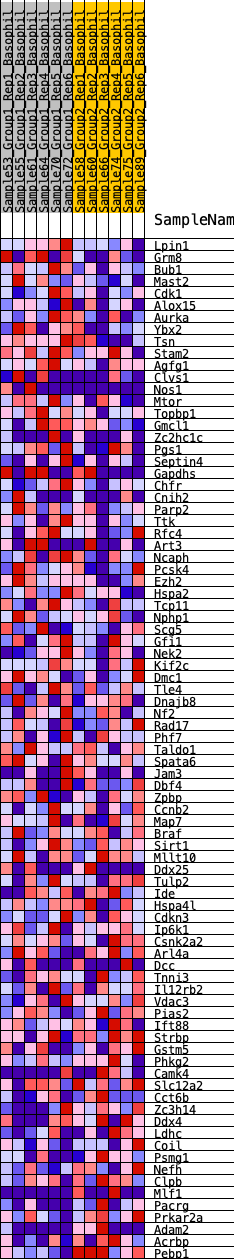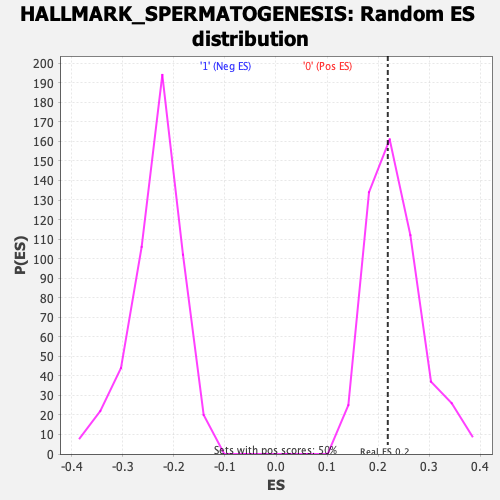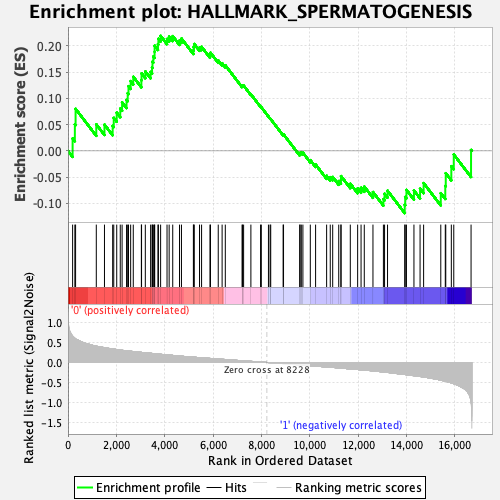
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.21904248 |
| Normalized Enrichment Score (NES) | 0.94571525 |
| Nominal p-value | 0.5416667 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lpin1 | 191 | 0.654 | 0.0237 | Yes |
| 2 | Grm8 | 284 | 0.599 | 0.0504 | Yes |
| 3 | Bub1 | 314 | 0.584 | 0.0801 | Yes |
| 4 | Mast2 | 1170 | 0.407 | 0.0505 | Yes |
| 5 | Cdk1 | 1508 | 0.368 | 0.0500 | Yes |
| 6 | Alox15 | 1847 | 0.336 | 0.0477 | Yes |
| 7 | Aurka | 1892 | 0.331 | 0.0629 | Yes |
| 8 | Ybx2 | 2015 | 0.321 | 0.0728 | Yes |
| 9 | Tsn | 2161 | 0.309 | 0.0807 | Yes |
| 10 | Stam2 | 2237 | 0.303 | 0.0925 | Yes |
| 11 | Agfg1 | 2421 | 0.290 | 0.0971 | Yes |
| 12 | Clvs1 | 2469 | 0.287 | 0.1098 | Yes |
| 13 | Nos1 | 2502 | 0.285 | 0.1232 | Yes |
| 14 | Mtor | 2591 | 0.280 | 0.1329 | Yes |
| 15 | Topbp1 | 2699 | 0.272 | 0.1411 | Yes |
| 16 | Gmcl1 | 3034 | 0.251 | 0.1345 | Yes |
| 17 | Zc2hc1c | 3042 | 0.251 | 0.1476 | Yes |
| 18 | Pgs1 | 3197 | 0.241 | 0.1513 | Yes |
| 19 | Septin4 | 3417 | 0.229 | 0.1504 | Yes |
| 20 | Gapdhs | 3476 | 0.225 | 0.1591 | Yes |
| 21 | Chfr | 3494 | 0.224 | 0.1701 | Yes |
| 22 | Cnih2 | 3529 | 0.221 | 0.1799 | Yes |
| 23 | Parp2 | 3580 | 0.218 | 0.1887 | Yes |
| 24 | Ttk | 3583 | 0.218 | 0.2003 | Yes |
| 25 | Rfc4 | 3721 | 0.209 | 0.2033 | Yes |
| 26 | Art3 | 3738 | 0.209 | 0.2135 | Yes |
| 27 | Ncaph | 3830 | 0.204 | 0.2190 | Yes |
| 28 | Pcsk4 | 4095 | 0.189 | 0.2133 | No |
| 29 | Ezh2 | 4183 | 0.184 | 0.2180 | No |
| 30 | Hspa2 | 4331 | 0.176 | 0.2186 | No |
| 31 | Tcp11 | 4612 | 0.161 | 0.2104 | No |
| 32 | Nphp1 | 4692 | 0.156 | 0.2140 | No |
| 33 | Scg5 | 5185 | 0.132 | 0.1915 | No |
| 34 | Gfi1 | 5198 | 0.132 | 0.1979 | No |
| 35 | Nek2 | 5220 | 0.130 | 0.2036 | No |
| 36 | Kif2c | 5439 | 0.120 | 0.1970 | No |
| 37 | Dmc1 | 5524 | 0.116 | 0.1982 | No |
| 38 | Tle4 | 5871 | 0.101 | 0.1828 | No |
| 39 | Dnajb8 | 5892 | 0.100 | 0.1869 | No |
| 40 | Nf2 | 6213 | 0.086 | 0.1723 | No |
| 41 | Rad17 | 6373 | 0.079 | 0.1670 | No |
| 42 | Phf7 | 6505 | 0.074 | 0.1630 | No |
| 43 | Taldo1 | 7198 | 0.043 | 0.1237 | No |
| 44 | Spata6 | 7217 | 0.042 | 0.1248 | No |
| 45 | Jam3 | 7262 | 0.040 | 0.1244 | No |
| 46 | Dbf4 | 7563 | 0.028 | 0.1078 | No |
| 47 | Zpbp | 7963 | 0.011 | 0.0844 | No |
| 48 | Ccnb2 | 7994 | 0.010 | 0.0831 | No |
| 49 | Map7 | 8289 | -0.001 | 0.0654 | No |
| 50 | Braf | 8356 | -0.004 | 0.0617 | No |
| 51 | Sirt1 | 8383 | -0.005 | 0.0604 | No |
| 52 | Mllt10 | 8896 | -0.026 | 0.0310 | No |
| 53 | Ddx25 | 8913 | -0.027 | 0.0314 | No |
| 54 | Tulp2 | 9581 | -0.058 | -0.0056 | No |
| 55 | Ide | 9599 | -0.059 | -0.0034 | No |
| 56 | Hspa4l | 9648 | -0.061 | -0.0030 | No |
| 57 | Cdkn3 | 9714 | -0.064 | -0.0035 | No |
| 58 | Ip6k1 | 10020 | -0.079 | -0.0176 | No |
| 59 | Csnk2a2 | 10239 | -0.089 | -0.0260 | No |
| 60 | Arl4a | 10695 | -0.110 | -0.0474 | No |
| 61 | Dcc | 10845 | -0.117 | -0.0501 | No |
| 62 | Tnni3 | 10946 | -0.120 | -0.0497 | No |
| 63 | Il12rb2 | 11196 | -0.135 | -0.0574 | No |
| 64 | Vdac3 | 11287 | -0.139 | -0.0553 | No |
| 65 | Pias2 | 11290 | -0.139 | -0.0480 | No |
| 66 | Ift88 | 11674 | -0.159 | -0.0625 | No |
| 67 | Strbp | 11978 | -0.173 | -0.0714 | No |
| 68 | Gstm5 | 12119 | -0.181 | -0.0701 | No |
| 69 | Phkg2 | 12252 | -0.188 | -0.0679 | No |
| 70 | Camk4 | 12611 | -0.207 | -0.0783 | No |
| 71 | Slc12a2 | 13043 | -0.234 | -0.0917 | No |
| 72 | Cct6b | 13091 | -0.238 | -0.0817 | No |
| 73 | Zc3h14 | 13214 | -0.246 | -0.0758 | No |
| 74 | Ddx4 | 13925 | -0.298 | -0.1025 | No |
| 75 | Ldhc | 13945 | -0.299 | -0.0876 | No |
| 76 | Coil | 13996 | -0.303 | -0.0743 | No |
| 77 | Psmg1 | 14306 | -0.326 | -0.0753 | No |
| 78 | Nefh | 14559 | -0.347 | -0.0718 | No |
| 79 | Clpb | 14706 | -0.359 | -0.0613 | No |
| 80 | Mlf1 | 15416 | -0.436 | -0.0805 | No |
| 81 | Pacrg | 15602 | -0.466 | -0.0666 | No |
| 82 | Prkar2a | 15622 | -0.469 | -0.0425 | No |
| 83 | Adam2 | 15852 | -0.505 | -0.0291 | No |
| 84 | Acrbp | 15953 | -0.525 | -0.0069 | No |
| 85 | Pebp1 | 16668 | -0.961 | 0.0019 | No |