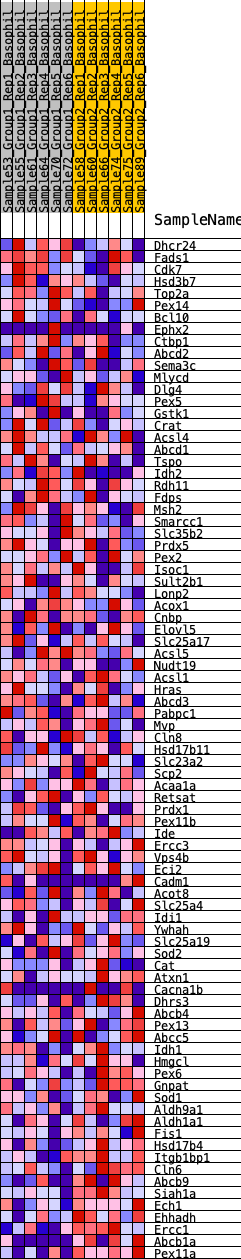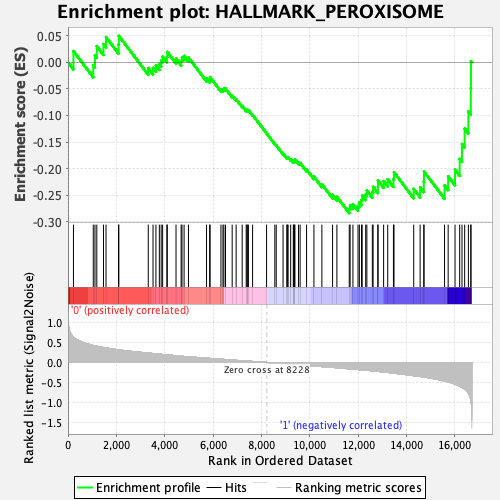
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.28368065 |
| Normalized Enrichment Score (NES) | -1.2234771 |
| Nominal p-value | 0.1554054 |
| FDR q-value | 0.7596381 |
| FWER p-Value | 0.927 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 225 | 0.636 | 0.0210 | No |
| 2 | Fads1 | 1045 | 0.423 | -0.0053 | No |
| 3 | Cdk7 | 1115 | 0.413 | 0.0130 | No |
| 4 | Hsd3b7 | 1191 | 0.403 | 0.0304 | No |
| 5 | Top2a | 1467 | 0.372 | 0.0341 | No |
| 6 | Pex14 | 1572 | 0.360 | 0.0473 | No |
| 7 | Bcl10 | 2090 | 0.314 | 0.0333 | No |
| 8 | Ephx2 | 2102 | 0.313 | 0.0496 | No |
| 9 | Ctbp1 | 3319 | 0.234 | -0.0108 | No |
| 10 | Abcd2 | 3516 | 0.222 | -0.0106 | No |
| 11 | Sema3c | 3636 | 0.214 | -0.0061 | No |
| 12 | Mlycd | 3778 | 0.207 | -0.0033 | No |
| 13 | Dlg4 | 3853 | 0.202 | 0.0032 | No |
| 14 | Pex5 | 3913 | 0.199 | 0.0105 | No |
| 15 | Gstk1 | 4090 | 0.189 | 0.0101 | No |
| 16 | Crat | 4105 | 0.188 | 0.0195 | No |
| 17 | Acsl4 | 4467 | 0.168 | 0.0069 | No |
| 18 | Abcd1 | 4678 | 0.157 | 0.0027 | No |
| 19 | Tspo | 4714 | 0.155 | 0.0090 | No |
| 20 | Idh2 | 4801 | 0.150 | 0.0120 | No |
| 21 | Rdh11 | 4981 | 0.141 | 0.0089 | No |
| 22 | Fdps | 5729 | 0.107 | -0.0302 | No |
| 23 | Msh2 | 5859 | 0.101 | -0.0325 | No |
| 24 | Smarcc1 | 5875 | 0.101 | -0.0279 | No |
| 25 | Slc35b2 | 6325 | 0.081 | -0.0505 | No |
| 26 | Prdx5 | 6408 | 0.077 | -0.0513 | No |
| 27 | Pex2 | 6447 | 0.076 | -0.0494 | No |
| 28 | Isoc1 | 6514 | 0.073 | -0.0494 | No |
| 29 | Sult2b1 | 6789 | 0.061 | -0.0626 | No |
| 30 | Lonp2 | 6953 | 0.054 | -0.0695 | No |
| 31 | Acox1 | 7203 | 0.043 | -0.0822 | No |
| 32 | Cnbp | 7369 | 0.036 | -0.0901 | No |
| 33 | Elovl5 | 7404 | 0.035 | -0.0903 | No |
| 34 | Slc25a17 | 7413 | 0.034 | -0.0889 | No |
| 35 | Acsl5 | 7461 | 0.032 | -0.0900 | No |
| 36 | Nudt19 | 7634 | 0.025 | -0.0990 | No |
| 37 | Acsl1 | 8212 | 0.001 | -0.1337 | No |
| 38 | Hras | 8553 | -0.012 | -0.1535 | No |
| 39 | Abcd3 | 8611 | -0.014 | -0.1561 | No |
| 40 | Pabpc1 | 8893 | -0.026 | -0.1716 | No |
| 41 | Mvp | 9048 | -0.033 | -0.1791 | No |
| 42 | Cln8 | 9072 | -0.034 | -0.1787 | No |
| 43 | Hsd17b11 | 9102 | -0.035 | -0.1785 | No |
| 44 | Slc23a2 | 9208 | -0.040 | -0.1826 | No |
| 45 | Scp2 | 9317 | -0.045 | -0.1867 | No |
| 46 | Acaa1a | 9334 | -0.046 | -0.1851 | No |
| 47 | Retsat | 9370 | -0.048 | -0.1847 | No |
| 48 | Prdx1 | 9382 | -0.048 | -0.1827 | No |
| 49 | Pex11b | 9533 | -0.056 | -0.1887 | No |
| 50 | Ide | 9599 | -0.059 | -0.1894 | No |
| 51 | Ercc3 | 9866 | -0.072 | -0.2015 | No |
| 52 | Vps4b | 10170 | -0.086 | -0.2151 | No |
| 53 | Eci2 | 10499 | -0.101 | -0.2293 | No |
| 54 | Cadm1 | 10941 | -0.120 | -0.2493 | No |
| 55 | Acot8 | 11120 | -0.130 | -0.2530 | No |
| 56 | Slc25a4 | 11631 | -0.156 | -0.2752 | Yes |
| 57 | Idi1 | 11666 | -0.158 | -0.2686 | Yes |
| 58 | Ywhah | 11783 | -0.164 | -0.2667 | Yes |
| 59 | Slc25a19 | 11993 | -0.174 | -0.2698 | Yes |
| 60 | Sod2 | 12049 | -0.177 | -0.2635 | Yes |
| 61 | Cat | 12138 | -0.182 | -0.2589 | Yes |
| 62 | Atxn1 | 12165 | -0.184 | -0.2505 | Yes |
| 63 | Cacna1b | 12311 | -0.192 | -0.2488 | Yes |
| 64 | Dhrs3 | 12359 | -0.194 | -0.2411 | Yes |
| 65 | Abcb4 | 12586 | -0.206 | -0.2435 | Yes |
| 66 | Pex13 | 12617 | -0.207 | -0.2341 | Yes |
| 67 | Abcc5 | 12816 | -0.221 | -0.2340 | Yes |
| 68 | Idh1 | 12819 | -0.221 | -0.2221 | Yes |
| 69 | Hmgcl | 13052 | -0.235 | -0.2233 | Yes |
| 70 | Pex6 | 13222 | -0.247 | -0.2201 | Yes |
| 71 | Gnpat | 13458 | -0.264 | -0.2199 | Yes |
| 72 | Sod1 | 13481 | -0.266 | -0.2068 | Yes |
| 73 | Aldh9a1 | 14297 | -0.326 | -0.2381 | Yes |
| 74 | Aldh1a1 | 14561 | -0.347 | -0.2351 | Yes |
| 75 | Fis1 | 14713 | -0.360 | -0.2246 | Yes |
| 76 | Hsd17b4 | 14723 | -0.361 | -0.2056 | Yes |
| 77 | Itgb1bp1 | 15572 | -0.462 | -0.2315 | Yes |
| 78 | Cln6 | 15721 | -0.481 | -0.2143 | Yes |
| 79 | Abcb9 | 16005 | -0.539 | -0.2020 | Yes |
| 80 | Siah1a | 16197 | -0.585 | -0.1817 | Yes |
| 81 | Ech1 | 16293 | -0.620 | -0.1538 | Yes |
| 82 | Ehhadh | 16403 | -0.658 | -0.1246 | Yes |
| 83 | Ercc1 | 16557 | -0.763 | -0.0924 | Yes |
| 84 | Abcb1a | 16658 | -0.917 | -0.0486 | Yes |
| 85 | Pex11a | 16664 | -0.938 | 0.0021 | Yes |