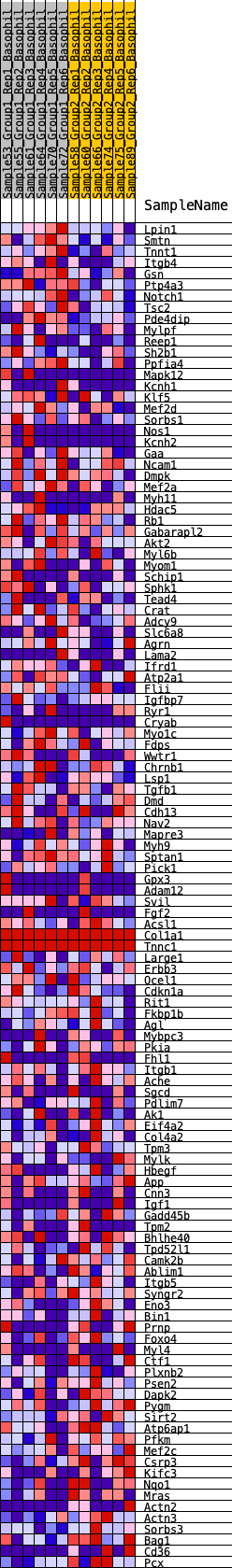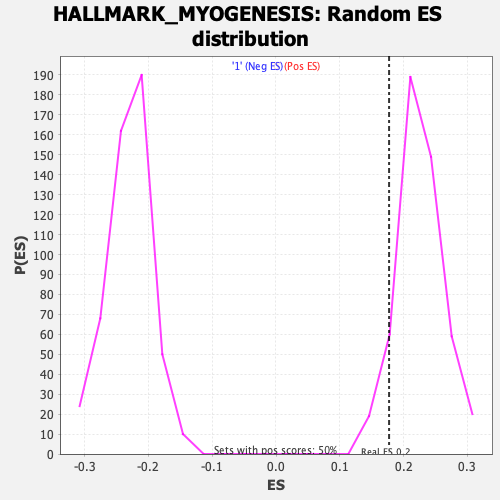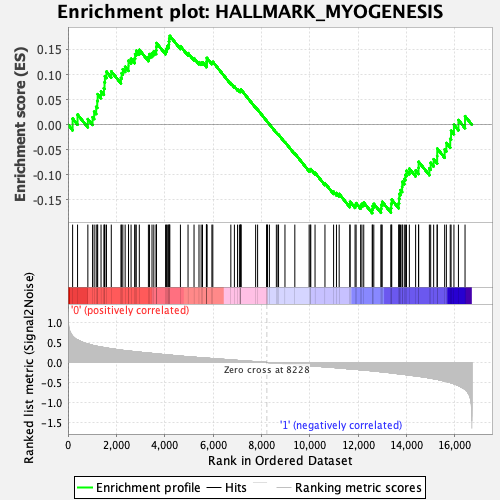
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.17716192 |
| Normalized Enrichment Score (NES) | 0.78695035 |
| Nominal p-value | 0.92741936 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lpin1 | 191 | 0.654 | 0.0121 | Yes |
| 2 | Smtn | 395 | 0.561 | 0.0201 | Yes |
| 3 | Tnnt1 | 821 | 0.461 | 0.0111 | Yes |
| 4 | Itgb4 | 1016 | 0.427 | 0.0148 | Yes |
| 5 | Gsn | 1081 | 0.418 | 0.0260 | Yes |
| 6 | Ptp4a3 | 1165 | 0.408 | 0.0357 | Yes |
| 7 | Notch1 | 1209 | 0.400 | 0.0476 | Yes |
| 8 | Tsc2 | 1228 | 0.398 | 0.0609 | Yes |
| 9 | Pde4dip | 1366 | 0.385 | 0.0665 | Yes |
| 10 | Mylpf | 1485 | 0.370 | 0.0727 | Yes |
| 11 | Reep1 | 1511 | 0.367 | 0.0845 | Yes |
| 12 | Sh2b1 | 1527 | 0.365 | 0.0968 | Yes |
| 13 | Ppfia4 | 1591 | 0.358 | 0.1059 | Yes |
| 14 | Mapk12 | 1790 | 0.341 | 0.1063 | Yes |
| 15 | Kcnh1 | 2190 | 0.307 | 0.0933 | Yes |
| 16 | Klf5 | 2219 | 0.305 | 0.1026 | Yes |
| 17 | Mef2d | 2268 | 0.301 | 0.1106 | Yes |
| 18 | Sorbs1 | 2363 | 0.294 | 0.1155 | Yes |
| 19 | Nos1 | 2502 | 0.285 | 0.1175 | Yes |
| 20 | Kcnh2 | 2503 | 0.285 | 0.1278 | Yes |
| 21 | Gaa | 2608 | 0.278 | 0.1316 | Yes |
| 22 | Ncam1 | 2758 | 0.269 | 0.1323 | Yes |
| 23 | Dmpk | 2775 | 0.268 | 0.1410 | Yes |
| 24 | Mef2a | 2823 | 0.263 | 0.1477 | Yes |
| 25 | Myh11 | 2949 | 0.256 | 0.1494 | Yes |
| 26 | Hdac5 | 3325 | 0.234 | 0.1352 | Yes |
| 27 | Rb1 | 3372 | 0.232 | 0.1408 | Yes |
| 28 | Gabarapl2 | 3474 | 0.225 | 0.1428 | Yes |
| 29 | Akt2 | 3549 | 0.220 | 0.1463 | Yes |
| 30 | Myl6b | 3644 | 0.214 | 0.1483 | Yes |
| 31 | Myom1 | 3652 | 0.213 | 0.1556 | Yes |
| 32 | Schip1 | 3660 | 0.213 | 0.1629 | Yes |
| 33 | Sphk1 | 4035 | 0.192 | 0.1473 | Yes |
| 34 | Tead4 | 4076 | 0.190 | 0.1517 | Yes |
| 35 | Crat | 4105 | 0.188 | 0.1568 | Yes |
| 36 | Adcy9 | 4167 | 0.185 | 0.1598 | Yes |
| 37 | Slc6a8 | 4174 | 0.184 | 0.1661 | Yes |
| 38 | Agrn | 4186 | 0.184 | 0.1720 | Yes |
| 39 | Lama2 | 4211 | 0.183 | 0.1772 | Yes |
| 40 | Ifrd1 | 4649 | 0.159 | 0.1565 | No |
| 41 | Atp2a1 | 4964 | 0.142 | 0.1427 | No |
| 42 | Flii | 5212 | 0.131 | 0.1326 | No |
| 43 | Igfbp7 | 5417 | 0.122 | 0.1246 | No |
| 44 | Ryr1 | 5506 | 0.117 | 0.1236 | No |
| 45 | Cryab | 5562 | 0.115 | 0.1244 | No |
| 46 | Myo1c | 5723 | 0.108 | 0.1186 | No |
| 47 | Fdps | 5729 | 0.107 | 0.1222 | No |
| 48 | Wwtr1 | 5734 | 0.107 | 0.1258 | No |
| 49 | Chrnb1 | 5740 | 0.107 | 0.1294 | No |
| 50 | Lsp1 | 5741 | 0.107 | 0.1332 | No |
| 51 | Tgfb1 | 5951 | 0.098 | 0.1241 | No |
| 52 | Dmd | 5989 | 0.096 | 0.1254 | No |
| 53 | Cdh13 | 6734 | 0.064 | 0.0828 | No |
| 54 | Nav2 | 6881 | 0.057 | 0.0761 | No |
| 55 | Mapre3 | 7011 | 0.051 | 0.0701 | No |
| 56 | Myh9 | 7088 | 0.047 | 0.0672 | No |
| 57 | Sptan1 | 7127 | 0.045 | 0.0666 | No |
| 58 | Pick1 | 7146 | 0.045 | 0.0671 | No |
| 59 | Gpx3 | 7149 | 0.045 | 0.0686 | No |
| 60 | Adam12 | 7150 | 0.045 | 0.0702 | No |
| 61 | Svil | 7759 | 0.019 | 0.0342 | No |
| 62 | Fgf2 | 7841 | 0.016 | 0.0299 | No |
| 63 | Acsl1 | 8212 | 0.001 | 0.0076 | No |
| 64 | Col1a1 | 8229 | 0.000 | 0.0067 | No |
| 65 | Tnnc1 | 8251 | 0.000 | 0.0054 | No |
| 66 | Large1 | 8329 | -0.002 | 0.0008 | No |
| 67 | Erbb3 | 8616 | -0.015 | -0.0159 | No |
| 68 | Ocel1 | 8681 | -0.018 | -0.0191 | No |
| 69 | Cdkn1a | 8707 | -0.018 | -0.0200 | No |
| 70 | Rit1 | 8972 | -0.029 | -0.0348 | No |
| 71 | Fkbp1b | 9380 | -0.048 | -0.0576 | No |
| 72 | Agl | 9972 | -0.077 | -0.0905 | No |
| 73 | Mybpc3 | 10019 | -0.079 | -0.0904 | No |
| 74 | Pkia | 10040 | -0.080 | -0.0888 | No |
| 75 | Fhl1 | 10216 | -0.088 | -0.0962 | No |
| 76 | Itgb1 | 10626 | -0.107 | -0.1170 | No |
| 77 | Ache | 10983 | -0.123 | -0.1340 | No |
| 78 | Sgcd | 11102 | -0.129 | -0.1365 | No |
| 79 | Pdlim7 | 11214 | -0.135 | -0.1383 | No |
| 80 | Ak1 | 11655 | -0.158 | -0.1591 | No |
| 81 | Eif4a2 | 11665 | -0.158 | -0.1540 | No |
| 82 | Col4a2 | 11871 | -0.168 | -0.1602 | No |
| 83 | Tpm3 | 11914 | -0.169 | -0.1567 | No |
| 84 | Mylk | 12101 | -0.181 | -0.1614 | No |
| 85 | Hbegf | 12155 | -0.183 | -0.1579 | No |
| 86 | App | 12230 | -0.187 | -0.1557 | No |
| 87 | Cnn3 | 12580 | -0.206 | -0.1693 | No |
| 88 | Igf1 | 12596 | -0.206 | -0.1627 | No |
| 89 | Gadd45b | 12646 | -0.209 | -0.1582 | No |
| 90 | Tpm2 | 12943 | -0.229 | -0.1677 | No |
| 91 | Bhlhe40 | 12954 | -0.230 | -0.1600 | No |
| 92 | Tpd52l1 | 12993 | -0.232 | -0.1540 | No |
| 93 | Camk2b | 13354 | -0.256 | -0.1664 | No |
| 94 | Ablim1 | 13365 | -0.257 | -0.1578 | No |
| 95 | Itgb5 | 13388 | -0.259 | -0.1497 | No |
| 96 | Syngr2 | 13677 | -0.279 | -0.1570 | No |
| 97 | Eno3 | 13691 | -0.280 | -0.1477 | No |
| 98 | Bin1 | 13703 | -0.281 | -0.1383 | No |
| 99 | Prnp | 13749 | -0.284 | -0.1307 | No |
| 100 | Foxo4 | 13817 | -0.290 | -0.1243 | No |
| 101 | Myl4 | 13822 | -0.290 | -0.1141 | No |
| 102 | Ctf1 | 13906 | -0.296 | -0.1084 | No |
| 103 | Plxnb2 | 13951 | -0.300 | -0.1002 | No |
| 104 | Psen2 | 13998 | -0.303 | -0.0921 | No |
| 105 | Dapk2 | 14116 | -0.312 | -0.0879 | No |
| 106 | Pygm | 14375 | -0.332 | -0.0914 | No |
| 107 | Sirt2 | 14496 | -0.342 | -0.0864 | No |
| 108 | Atp6ap1 | 14501 | -0.342 | -0.0743 | No |
| 109 | Pfkm | 14944 | -0.383 | -0.0871 | No |
| 110 | Mef2c | 14998 | -0.390 | -0.0762 | No |
| 111 | Csrp3 | 15121 | -0.402 | -0.0691 | No |
| 112 | Kifc3 | 15266 | -0.417 | -0.0627 | No |
| 113 | Nqo1 | 15270 | -0.418 | -0.0478 | No |
| 114 | Mras | 15576 | -0.463 | -0.0495 | No |
| 115 | Actn2 | 15647 | -0.473 | -0.0367 | No |
| 116 | Actn3 | 15806 | -0.497 | -0.0282 | No |
| 117 | Sorbs3 | 15841 | -0.504 | -0.0121 | No |
| 118 | Bag1 | 15960 | -0.527 | -0.0002 | No |
| 119 | Cd36 | 16146 | -0.570 | 0.0092 | No |
| 120 | Pcx | 16419 | -0.668 | 0.0169 | No |