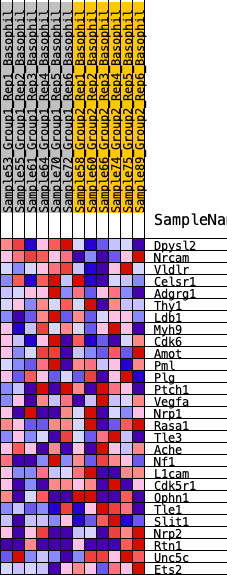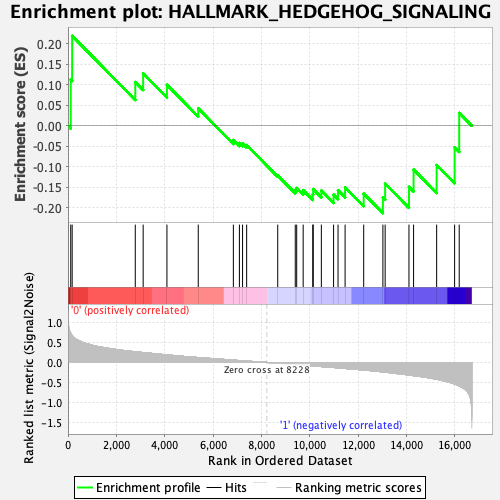
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.21920352 |
| Normalized Enrichment Score (NES) | 0.7252187 |
| Nominal p-value | 0.8772277 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dpysl2 | 113 | 0.726 | 0.1126 | Yes |
| 2 | Nrcam | 172 | 0.670 | 0.2192 | Yes |
| 3 | Vldlr | 2782 | 0.267 | 0.1066 | No |
| 4 | Celsr1 | 3108 | 0.247 | 0.1277 | No |
| 5 | Adgrg1 | 4091 | 0.189 | 0.0998 | No |
| 6 | Thy1 | 5386 | 0.123 | 0.0424 | No |
| 7 | Ldb1 | 6838 | 0.059 | -0.0348 | No |
| 8 | Myh9 | 7088 | 0.047 | -0.0421 | No |
| 9 | Cdk6 | 7219 | 0.042 | -0.0430 | No |
| 10 | Amot | 7388 | 0.035 | -0.0473 | No |
| 11 | Pml | 8673 | -0.017 | -0.1214 | No |
| 12 | Plg | 9398 | -0.049 | -0.1568 | No |
| 13 | Ptch1 | 9457 | -0.052 | -0.1518 | No |
| 14 | Vegfa | 9725 | -0.064 | -0.1573 | No |
| 15 | Nrp1 | 10122 | -0.084 | -0.1673 | No |
| 16 | Rasa1 | 10141 | -0.084 | -0.1545 | No |
| 17 | Tle3 | 10477 | -0.101 | -0.1581 | No |
| 18 | Ache | 10983 | -0.123 | -0.1682 | No |
| 19 | Nf1 | 11167 | -0.133 | -0.1572 | No |
| 20 | L1cam | 11459 | -0.148 | -0.1503 | No |
| 21 | Cdk5r1 | 12228 | -0.187 | -0.1656 | No |
| 22 | Ophn1 | 13022 | -0.233 | -0.1749 | No |
| 23 | Tle1 | 13115 | -0.239 | -0.1411 | No |
| 24 | Slit1 | 14100 | -0.311 | -0.1490 | No |
| 25 | Nrp2 | 14291 | -0.326 | -0.1069 | No |
| 26 | Rtn1 | 15245 | -0.414 | -0.0960 | No |
| 27 | Unc5c | 15988 | -0.534 | -0.0527 | No |
| 28 | Ets2 | 16178 | -0.580 | 0.0313 | No |