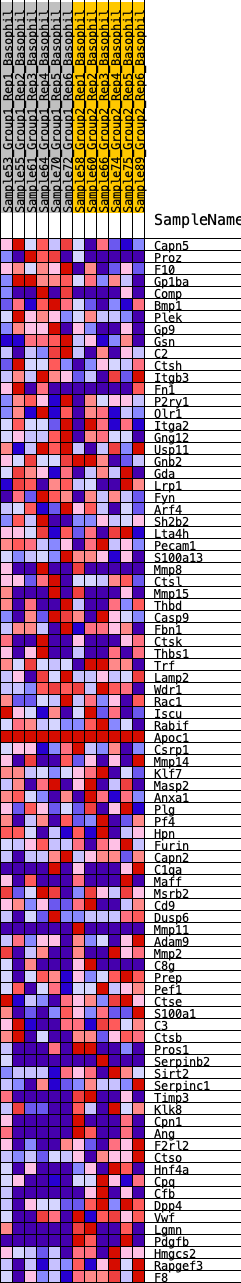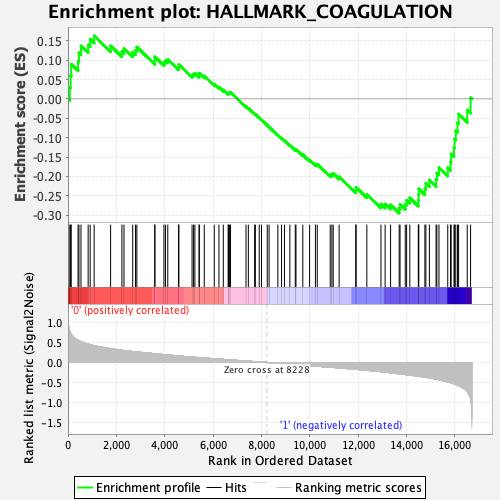
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.29456186 |
| Normalized Enrichment Score (NES) | -1.261668 |
| Nominal p-value | 0.09677419 |
| FDR q-value | 0.709755 |
| FWER p-Value | 0.89 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Capn5 | 75 | 0.773 | 0.0297 | No |
| 2 | Proz | 108 | 0.732 | 0.0601 | No |
| 3 | F10 | 132 | 0.700 | 0.0896 | No |
| 4 | Gp1ba | 417 | 0.552 | 0.0969 | No |
| 5 | Comp | 452 | 0.543 | 0.1189 | No |
| 6 | Bmp1 | 537 | 0.519 | 0.1367 | No |
| 7 | Plek | 833 | 0.459 | 0.1392 | No |
| 8 | Gp9 | 917 | 0.445 | 0.1539 | No |
| 9 | Gsn | 1081 | 0.418 | 0.1626 | No |
| 10 | C2 | 1762 | 0.344 | 0.1369 | No |
| 11 | Ctsh | 2229 | 0.304 | 0.1222 | No |
| 12 | Itgb3 | 2315 | 0.297 | 0.1303 | No |
| 13 | Fn1 | 2676 | 0.274 | 0.1207 | No |
| 14 | P2ry1 | 2790 | 0.266 | 0.1257 | No |
| 15 | Olr1 | 2847 | 0.262 | 0.1339 | No |
| 16 | Itga2 | 3590 | 0.217 | 0.0988 | No |
| 17 | Gng12 | 3592 | 0.217 | 0.1084 | No |
| 18 | Usp11 | 3967 | 0.196 | 0.0945 | No |
| 19 | Gnb2 | 4030 | 0.192 | 0.0993 | No |
| 20 | Gda | 4126 | 0.187 | 0.1018 | No |
| 21 | Lrp1 | 4570 | 0.163 | 0.0823 | No |
| 22 | Fyn | 4586 | 0.162 | 0.0886 | No |
| 23 | Arf4 | 5130 | 0.135 | 0.0618 | No |
| 24 | Sh2b2 | 5193 | 0.132 | 0.0639 | No |
| 25 | Lta4h | 5248 | 0.129 | 0.0664 | No |
| 26 | Pecam1 | 5420 | 0.122 | 0.0615 | No |
| 27 | S100a13 | 5429 | 0.121 | 0.0663 | No |
| 28 | Mmp8 | 5635 | 0.112 | 0.0589 | No |
| 29 | Ctsl | 6049 | 0.093 | 0.0382 | No |
| 30 | Mmp15 | 6242 | 0.084 | 0.0304 | No |
| 31 | Thbd | 6422 | 0.077 | 0.0230 | No |
| 32 | Casp9 | 6623 | 0.069 | 0.0140 | No |
| 33 | Fbn1 | 6638 | 0.068 | 0.0162 | No |
| 34 | Ctsk | 6674 | 0.067 | 0.0170 | No |
| 35 | Thbs1 | 6721 | 0.064 | 0.0171 | No |
| 36 | Trf | 7361 | 0.037 | -0.0198 | No |
| 37 | Lamp2 | 7458 | 0.032 | -0.0241 | No |
| 38 | Wdr1 | 7718 | 0.021 | -0.0388 | No |
| 39 | Rac1 | 7747 | 0.020 | -0.0396 | No |
| 40 | Iscu | 7908 | 0.014 | -0.0486 | No |
| 41 | Rabif | 8001 | 0.009 | -0.0538 | No |
| 42 | Apoc1 | 8240 | 0.000 | -0.0681 | No |
| 43 | Csrp1 | 8316 | -0.002 | -0.0725 | No |
| 44 | Mmp14 | 8678 | -0.017 | -0.0935 | No |
| 45 | Klf7 | 8831 | -0.023 | -0.1016 | No |
| 46 | Masp2 | 8951 | -0.028 | -0.1075 | No |
| 47 | Anxa1 | 9175 | -0.039 | -0.1192 | No |
| 48 | Plg | 9398 | -0.049 | -0.1304 | No |
| 49 | Pf4 | 9427 | -0.050 | -0.1299 | No |
| 50 | Hpn | 9710 | -0.064 | -0.1441 | No |
| 51 | Furin | 9991 | -0.077 | -0.1575 | No |
| 52 | Capn2 | 10231 | -0.088 | -0.1680 | No |
| 53 | C1qa | 10308 | -0.092 | -0.1685 | No |
| 54 | Maff | 10844 | -0.117 | -0.1955 | No |
| 55 | Msrb2 | 10912 | -0.119 | -0.1943 | No |
| 56 | Cd9 | 10968 | -0.122 | -0.1923 | No |
| 57 | Dusp6 | 11212 | -0.135 | -0.2009 | No |
| 58 | Mmp11 | 11902 | -0.169 | -0.2349 | No |
| 59 | Adam9 | 11915 | -0.169 | -0.2281 | No |
| 60 | Mmp2 | 12358 | -0.194 | -0.2462 | No |
| 61 | C8g | 12942 | -0.229 | -0.2712 | No |
| 62 | Prep | 13114 | -0.239 | -0.2709 | No |
| 63 | Pef1 | 13340 | -0.255 | -0.2732 | No |
| 64 | Ctse | 13696 | -0.280 | -0.2822 | Yes |
| 65 | S100a1 | 13731 | -0.283 | -0.2717 | Yes |
| 66 | C3 | 13946 | -0.299 | -0.2714 | Yes |
| 67 | Ctsb | 14003 | -0.303 | -0.2613 | Yes |
| 68 | Pros1 | 14135 | -0.314 | -0.2553 | Yes |
| 69 | Serpinb2 | 14487 | -0.341 | -0.2614 | Yes |
| 70 | Sirt2 | 14496 | -0.342 | -0.2468 | Yes |
| 71 | Serpinc1 | 14506 | -0.342 | -0.2322 | Yes |
| 72 | Timp3 | 14759 | -0.364 | -0.2313 | Yes |
| 73 | Klk8 | 14799 | -0.369 | -0.2174 | Yes |
| 74 | Cpn1 | 14945 | -0.383 | -0.2091 | Yes |
| 75 | Ang | 15221 | -0.412 | -0.2075 | Yes |
| 76 | F2rl2 | 15254 | -0.416 | -0.1911 | Yes |
| 77 | Ctso | 15341 | -0.429 | -0.1773 | Yes |
| 78 | Hnf4a | 15704 | -0.479 | -0.1779 | Yes |
| 79 | Cpq | 15818 | -0.500 | -0.1626 | Yes |
| 80 | Cfb | 15844 | -0.504 | -0.1419 | Yes |
| 81 | Dpp4 | 15963 | -0.527 | -0.1257 | Yes |
| 82 | Vwf | 15987 | -0.534 | -0.1035 | Yes |
| 83 | Lgmn | 16036 | -0.547 | -0.0822 | Yes |
| 84 | Pdgfb | 16110 | -0.562 | -0.0617 | Yes |
| 85 | Hmgcs2 | 16153 | -0.573 | -0.0390 | Yes |
| 86 | Rapgef3 | 16510 | -0.719 | -0.0286 | Yes |
| 87 | F8 | 16648 | -0.904 | 0.0031 | Yes |