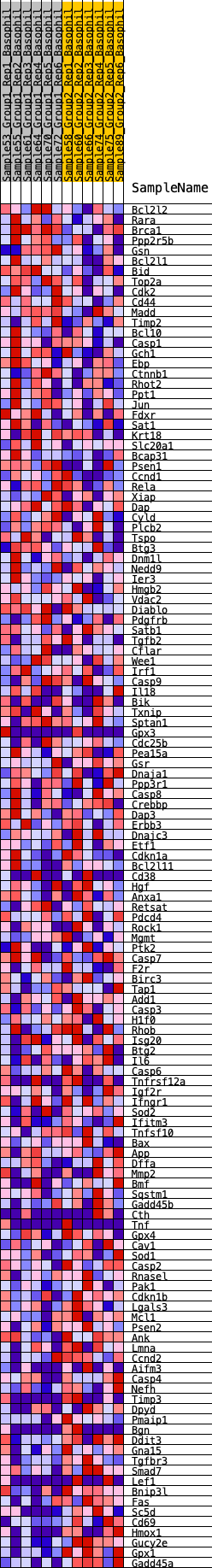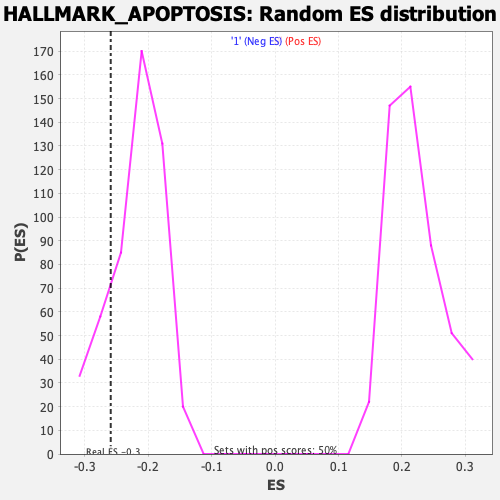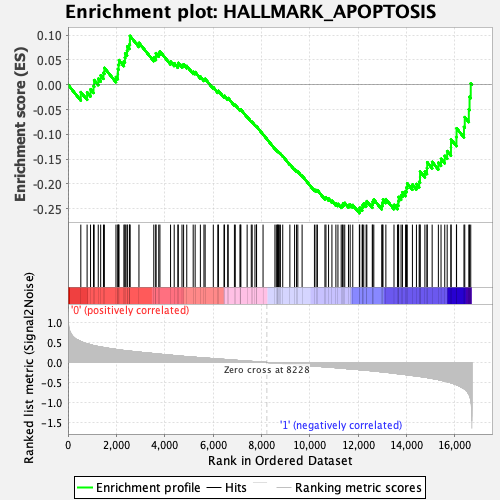
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.25918162 |
| Normalized Enrichment Score (NES) | -1.1814058 |
| Nominal p-value | 0.18309858 |
| FDR q-value | 0.68920237 |
| FWER p-Value | 0.957 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bcl2l2 | 530 | 0.522 | -0.0149 | No |
| 2 | Rara | 790 | 0.465 | -0.0153 | No |
| 3 | Brca1 | 932 | 0.442 | -0.0093 | No |
| 4 | Ppp2r5b | 1055 | 0.422 | -0.0029 | No |
| 5 | Gsn | 1081 | 0.418 | 0.0093 | No |
| 6 | Bcl2l1 | 1251 | 0.396 | 0.0121 | No |
| 7 | Bid | 1351 | 0.387 | 0.0188 | No |
| 8 | Top2a | 1467 | 0.372 | 0.0241 | No |
| 9 | Cdk2 | 1504 | 0.368 | 0.0340 | No |
| 10 | Cd44 | 1984 | 0.324 | 0.0157 | No |
| 11 | Madd | 2062 | 0.318 | 0.0214 | No |
| 12 | Timp2 | 2065 | 0.317 | 0.0317 | No |
| 13 | Bcl10 | 2090 | 0.314 | 0.0406 | No |
| 14 | Casp1 | 2109 | 0.313 | 0.0497 | No |
| 15 | Gch1 | 2312 | 0.297 | 0.0473 | No |
| 16 | Ebp | 2354 | 0.295 | 0.0545 | No |
| 17 | Ctnnb1 | 2365 | 0.294 | 0.0635 | No |
| 18 | Rhot2 | 2442 | 0.289 | 0.0684 | No |
| 19 | Ppt1 | 2451 | 0.288 | 0.0773 | No |
| 20 | Jun | 2539 | 0.283 | 0.0813 | No |
| 21 | Fdxr | 2560 | 0.281 | 0.0894 | No |
| 22 | Sat1 | 2561 | 0.281 | 0.0986 | No |
| 23 | Krt18 | 2934 | 0.256 | 0.0845 | No |
| 24 | Slc20a1 | 3548 | 0.220 | 0.0547 | No |
| 25 | Bcap31 | 3634 | 0.215 | 0.0566 | No |
| 26 | Psen1 | 3635 | 0.215 | 0.0637 | No |
| 27 | Ccnd1 | 3753 | 0.208 | 0.0634 | No |
| 28 | Rela | 3801 | 0.206 | 0.0673 | No |
| 29 | Xiap | 4241 | 0.181 | 0.0467 | No |
| 30 | Dap | 4392 | 0.172 | 0.0433 | No |
| 31 | Cyld | 4544 | 0.164 | 0.0396 | No |
| 32 | Plcb2 | 4560 | 0.163 | 0.0440 | No |
| 33 | Tspo | 4714 | 0.155 | 0.0399 | No |
| 34 | Btg3 | 4780 | 0.151 | 0.0409 | No |
| 35 | Dnm1l | 4911 | 0.145 | 0.0378 | No |
| 36 | Nedd9 | 5177 | 0.132 | 0.0261 | No |
| 37 | Ier3 | 5258 | 0.129 | 0.0255 | No |
| 38 | Hmgb2 | 5474 | 0.118 | 0.0164 | No |
| 39 | Vdac2 | 5622 | 0.113 | 0.0112 | No |
| 40 | Diablo | 5671 | 0.110 | 0.0120 | No |
| 41 | Pdgfrb | 6009 | 0.095 | -0.0053 | No |
| 42 | Satb1 | 6196 | 0.087 | -0.0136 | No |
| 43 | Tgfb2 | 6221 | 0.085 | -0.0123 | No |
| 44 | Cflar | 6451 | 0.076 | -0.0236 | No |
| 45 | Wee1 | 6473 | 0.075 | -0.0224 | No |
| 46 | Irf1 | 6606 | 0.069 | -0.0281 | No |
| 47 | Casp9 | 6623 | 0.069 | -0.0268 | No |
| 48 | Il18 | 6882 | 0.057 | -0.0405 | No |
| 49 | Bik | 6917 | 0.055 | -0.0408 | No |
| 50 | Txnip | 7114 | 0.046 | -0.0511 | No |
| 51 | Sptan1 | 7127 | 0.045 | -0.0504 | No |
| 52 | Gpx3 | 7149 | 0.045 | -0.0502 | No |
| 53 | Cdc25b | 7408 | 0.035 | -0.0646 | No |
| 54 | Pea15a | 7579 | 0.027 | -0.0740 | No |
| 55 | Gsr | 7616 | 0.026 | -0.0753 | No |
| 56 | Dnaja1 | 7731 | 0.020 | -0.0815 | No |
| 57 | Ppp3r1 | 7796 | 0.018 | -0.0848 | No |
| 58 | Casp8 | 7797 | 0.018 | -0.0842 | No |
| 59 | Crebbp | 8066 | 0.007 | -0.1001 | No |
| 60 | Dap3 | 8558 | -0.012 | -0.1294 | No |
| 61 | Erbb3 | 8616 | -0.015 | -0.1323 | No |
| 62 | Dnajc3 | 8662 | -0.017 | -0.1345 | No |
| 63 | Etf1 | 8677 | -0.017 | -0.1348 | No |
| 64 | Cdkn1a | 8707 | -0.018 | -0.1359 | No |
| 65 | Bcl2l11 | 8738 | -0.020 | -0.1371 | No |
| 66 | Cd38 | 8783 | -0.021 | -0.1391 | No |
| 67 | Hgf | 8880 | -0.026 | -0.1440 | No |
| 68 | Anxa1 | 9175 | -0.039 | -0.1605 | No |
| 69 | Retsat | 9370 | -0.048 | -0.1706 | No |
| 70 | Pdcd4 | 9451 | -0.051 | -0.1738 | No |
| 71 | Rock1 | 9504 | -0.054 | -0.1752 | No |
| 72 | Mgmt | 9681 | -0.062 | -0.1837 | No |
| 73 | Ptk2 | 10191 | -0.087 | -0.2116 | No |
| 74 | Casp7 | 10259 | -0.090 | -0.2127 | No |
| 75 | F2r | 10314 | -0.092 | -0.2130 | No |
| 76 | Birc3 | 10625 | -0.107 | -0.2282 | No |
| 77 | Tap1 | 10667 | -0.109 | -0.2271 | No |
| 78 | Add1 | 10775 | -0.115 | -0.2297 | No |
| 79 | Casp3 | 10914 | -0.119 | -0.2342 | No |
| 80 | H1f0 | 11078 | -0.128 | -0.2398 | No |
| 81 | Rhob | 11163 | -0.133 | -0.2405 | No |
| 82 | Isg20 | 11297 | -0.139 | -0.2440 | No |
| 83 | Btg2 | 11348 | -0.142 | -0.2424 | No |
| 84 | Il6 | 11378 | -0.144 | -0.2394 | No |
| 85 | Casp6 | 11444 | -0.147 | -0.2385 | No |
| 86 | Tnfrsf12a | 11599 | -0.155 | -0.2427 | No |
| 87 | Igf2r | 11668 | -0.158 | -0.2417 | No |
| 88 | Ifngr1 | 11776 | -0.164 | -0.2428 | No |
| 89 | Sod2 | 12049 | -0.177 | -0.2534 | Yes |
| 90 | Ifitm3 | 12063 | -0.178 | -0.2483 | Yes |
| 91 | Tnfsf10 | 12164 | -0.183 | -0.2483 | Yes |
| 92 | Bax | 12180 | -0.185 | -0.2432 | Yes |
| 93 | App | 12230 | -0.187 | -0.2400 | Yes |
| 94 | Dffa | 12329 | -0.192 | -0.2396 | Yes |
| 95 | Mmp2 | 12358 | -0.194 | -0.2350 | Yes |
| 96 | Bmf | 12583 | -0.206 | -0.2417 | Yes |
| 97 | Sqstm1 | 12600 | -0.206 | -0.2359 | Yes |
| 98 | Gadd45b | 12646 | -0.209 | -0.2318 | Yes |
| 99 | Cth | 12975 | -0.231 | -0.2440 | Yes |
| 100 | Tnf | 13001 | -0.232 | -0.2379 | Yes |
| 101 | Gpx4 | 13026 | -0.233 | -0.2317 | Yes |
| 102 | Cav1 | 13145 | -0.241 | -0.2310 | Yes |
| 103 | Sod1 | 13481 | -0.266 | -0.2425 | Yes |
| 104 | Casp2 | 13625 | -0.277 | -0.2420 | Yes |
| 105 | Rnasel | 13664 | -0.278 | -0.2352 | Yes |
| 106 | Pak1 | 13673 | -0.279 | -0.2265 | Yes |
| 107 | Cdkn1b | 13773 | -0.286 | -0.2232 | Yes |
| 108 | Lgals3 | 13830 | -0.291 | -0.2170 | Yes |
| 109 | Mcl1 | 13960 | -0.300 | -0.2150 | Yes |
| 110 | Psen2 | 13998 | -0.303 | -0.2073 | Yes |
| 111 | Ank | 14029 | -0.305 | -0.1991 | Yes |
| 112 | Lmna | 14243 | -0.323 | -0.2014 | Yes |
| 113 | Ccnd2 | 14409 | -0.335 | -0.2004 | Yes |
| 114 | Aifm3 | 14522 | -0.344 | -0.1958 | Yes |
| 115 | Casp4 | 14557 | -0.347 | -0.1865 | Yes |
| 116 | Nefh | 14559 | -0.347 | -0.1752 | Yes |
| 117 | Timp3 | 14759 | -0.364 | -0.1753 | Yes |
| 118 | Dpyd | 14845 | -0.373 | -0.1682 | Yes |
| 119 | Pmaip1 | 14855 | -0.374 | -0.1565 | Yes |
| 120 | Bgn | 15059 | -0.397 | -0.1557 | Yes |
| 121 | Ddit3 | 15316 | -0.424 | -0.1573 | Yes |
| 122 | Gna15 | 15426 | -0.437 | -0.1495 | Yes |
| 123 | Tgfbr3 | 15580 | -0.463 | -0.1436 | Yes |
| 124 | Smad7 | 15680 | -0.476 | -0.1340 | Yes |
| 125 | Lef1 | 15837 | -0.503 | -0.1269 | Yes |
| 126 | Bnip3l | 15840 | -0.504 | -0.1105 | Yes |
| 127 | Fas | 16063 | -0.553 | -0.1058 | Yes |
| 128 | Sc5d | 16069 | -0.554 | -0.0879 | Yes |
| 129 | Cd69 | 16375 | -0.648 | -0.0851 | Yes |
| 130 | Hmox1 | 16408 | -0.661 | -0.0654 | Yes |
| 131 | Gucy2e | 16576 | -0.783 | -0.0498 | Yes |
| 132 | Gpx1 | 16606 | -0.827 | -0.0244 | Yes |
| 133 | Gadd45a | 16655 | -0.915 | 0.0027 | Yes |