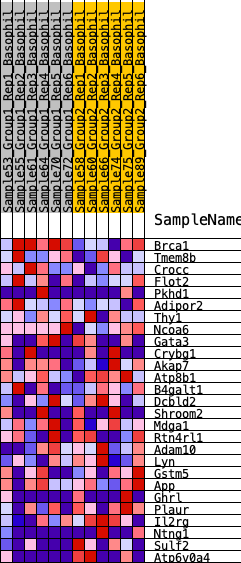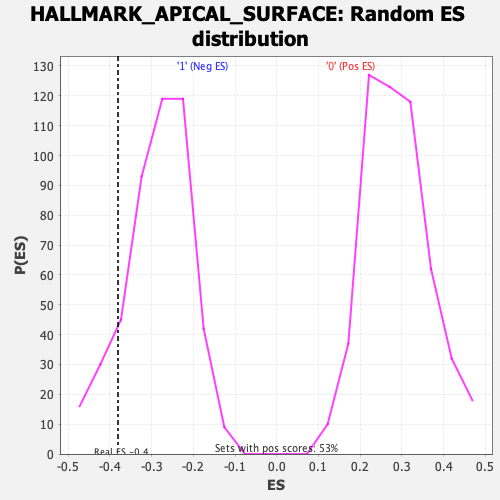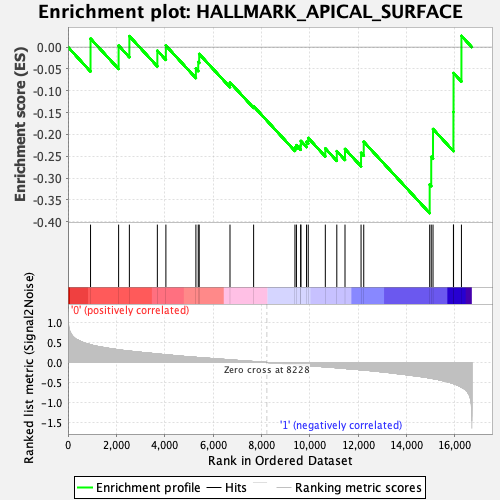
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | -0.3803774 |
| Normalized Enrichment Score (NES) | -1.3263085 |
| Nominal p-value | 0.12684989 |
| FDR q-value | 0.56911784 |
| FWER p-Value | 0.809 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Brca1 | 932 | 0.442 | 0.0195 | No |
| 2 | Tmem8b | 2093 | 0.314 | 0.0035 | No |
| 3 | Crocc | 2538 | 0.283 | 0.0252 | No |
| 4 | Flot2 | 3695 | 0.211 | -0.0082 | No |
| 5 | Pkhd1 | 4048 | 0.191 | 0.0034 | No |
| 6 | Adipor2 | 5289 | 0.127 | -0.0493 | No |
| 7 | Thy1 | 5386 | 0.123 | -0.0340 | No |
| 8 | Ncoa6 | 5426 | 0.121 | -0.0157 | No |
| 9 | Gata3 | 6699 | 0.065 | -0.0809 | No |
| 10 | Crybg1 | 7676 | 0.023 | -0.1355 | No |
| 11 | Akap7 | 9385 | -0.048 | -0.2297 | No |
| 12 | Atp8b1 | 9447 | -0.051 | -0.2247 | No |
| 13 | B4galt1 | 9624 | -0.060 | -0.2250 | No |
| 14 | Dcbld2 | 9629 | -0.060 | -0.2150 | No |
| 15 | Shroom2 | 9864 | -0.072 | -0.2168 | No |
| 16 | Mdga1 | 9935 | -0.075 | -0.2082 | No |
| 17 | Rtn4rl1 | 10641 | -0.108 | -0.2321 | No |
| 18 | Adam10 | 11116 | -0.130 | -0.2384 | No |
| 19 | Lyn | 11455 | -0.148 | -0.2334 | No |
| 20 | Gstm5 | 12119 | -0.181 | -0.2422 | No |
| 21 | App | 12230 | -0.187 | -0.2169 | No |
| 22 | Ghrl | 14957 | -0.384 | -0.3148 | Yes |
| 23 | Plaur | 15021 | -0.393 | -0.2515 | Yes |
| 24 | Il2rg | 15095 | -0.400 | -0.1876 | Yes |
| 25 | Ntng1 | 15939 | -0.524 | -0.1487 | Yes |
| 26 | Sulf2 | 15944 | -0.525 | -0.0595 | Yes |
| 27 | Atp6v0a4 | 16270 | -0.614 | 0.0257 | Yes |