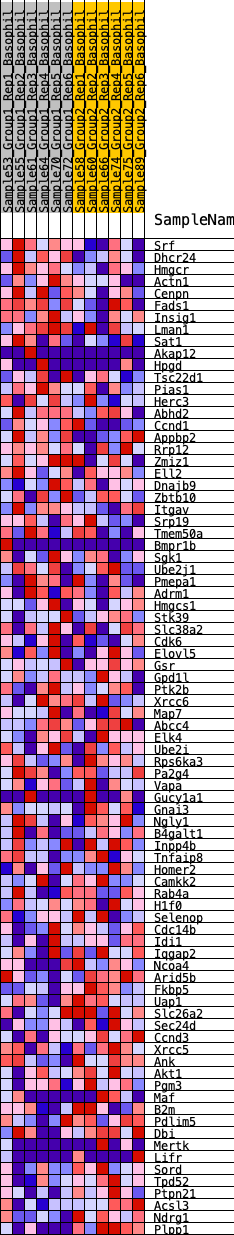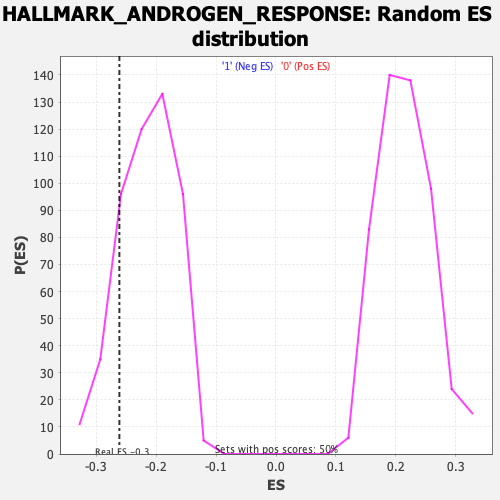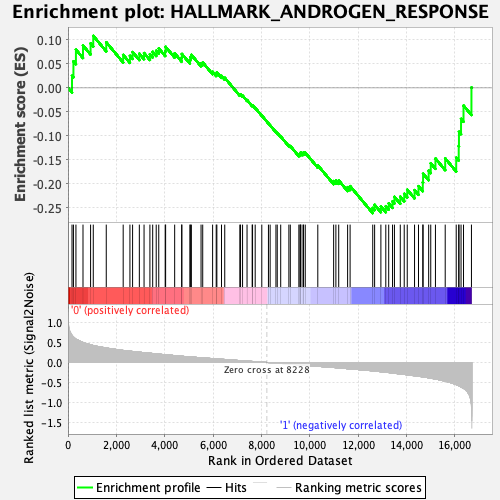
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ANDROGEN_RESPONSE |
| Enrichment Score (ES) | -0.2610883 |
| Normalized Enrichment Score (NES) | -1.2189606 |
| Nominal p-value | 0.15524194 |
| FDR q-value | 0.69230926 |
| FWER p-Value | 0.932 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Srf | 166 | 0.674 | 0.0255 | No |
| 2 | Dhcr24 | 225 | 0.636 | 0.0554 | No |
| 3 | Hmgcr | 328 | 0.578 | 0.0797 | No |
| 4 | Actn1 | 620 | 0.498 | 0.0884 | No |
| 5 | Cenpn | 934 | 0.442 | 0.0928 | No |
| 6 | Fads1 | 1045 | 0.423 | 0.1085 | No |
| 7 | Insig1 | 1582 | 0.359 | 0.0951 | No |
| 8 | Lman1 | 2282 | 0.300 | 0.0688 | No |
| 9 | Sat1 | 2561 | 0.281 | 0.0669 | No |
| 10 | Akap12 | 2671 | 0.274 | 0.0747 | No |
| 11 | Hpgd | 2950 | 0.256 | 0.0715 | No |
| 12 | Tsc22d1 | 3145 | 0.244 | 0.0726 | No |
| 13 | Pias1 | 3387 | 0.231 | 0.0703 | No |
| 14 | Herc3 | 3502 | 0.223 | 0.0752 | No |
| 15 | Abhd2 | 3648 | 0.214 | 0.0777 | No |
| 16 | Ccnd1 | 3753 | 0.208 | 0.0823 | No |
| 17 | Appbp2 | 4019 | 0.193 | 0.0765 | No |
| 18 | Rrp12 | 4037 | 0.192 | 0.0856 | No |
| 19 | Zmiz1 | 4411 | 0.171 | 0.0722 | No |
| 20 | Ell2 | 4701 | 0.156 | 0.0630 | No |
| 21 | Dnajb9 | 4708 | 0.155 | 0.0708 | No |
| 22 | Zbtb10 | 5041 | 0.139 | 0.0581 | No |
| 23 | Itgav | 5061 | 0.138 | 0.0642 | No |
| 24 | Srp19 | 5102 | 0.136 | 0.0690 | No |
| 25 | Tmem50a | 5502 | 0.117 | 0.0511 | No |
| 26 | Bmpr1b | 5571 | 0.115 | 0.0530 | No |
| 27 | Sgk1 | 5980 | 0.096 | 0.0336 | No |
| 28 | Ube2j1 | 6129 | 0.089 | 0.0293 | No |
| 29 | Pmepa1 | 6156 | 0.088 | 0.0324 | No |
| 30 | Adrm1 | 6348 | 0.080 | 0.0251 | No |
| 31 | Hmgcs1 | 6482 | 0.075 | 0.0211 | No |
| 32 | Stk39 | 7106 | 0.046 | -0.0140 | No |
| 33 | Slc38a2 | 7135 | 0.045 | -0.0133 | No |
| 34 | Cdk6 | 7219 | 0.042 | -0.0161 | No |
| 35 | Elovl5 | 7404 | 0.035 | -0.0254 | No |
| 36 | Gsr | 7616 | 0.026 | -0.0367 | No |
| 37 | Gpd1l | 7632 | 0.025 | -0.0363 | No |
| 38 | Ptk2b | 7741 | 0.020 | -0.0418 | No |
| 39 | Xrcc6 | 8014 | 0.009 | -0.0577 | No |
| 40 | Map7 | 8289 | -0.001 | -0.0741 | No |
| 41 | Abcc4 | 8359 | -0.004 | -0.0780 | No |
| 42 | Elk4 | 8599 | -0.014 | -0.0917 | No |
| 43 | Ube2i | 8653 | -0.017 | -0.0940 | No |
| 44 | Rps6ka3 | 8796 | -0.022 | -0.1014 | No |
| 45 | Pa2g4 | 9134 | -0.037 | -0.1198 | No |
| 46 | Vapa | 9193 | -0.039 | -0.1212 | No |
| 47 | Gucy1a1 | 9547 | -0.057 | -0.1394 | No |
| 48 | Gnai3 | 9592 | -0.059 | -0.1390 | No |
| 49 | Ngly1 | 9623 | -0.060 | -0.1376 | No |
| 50 | B4galt1 | 9624 | -0.060 | -0.1345 | No |
| 51 | Inpp4b | 9719 | -0.064 | -0.1368 | No |
| 52 | Tnfaip8 | 9740 | -0.066 | -0.1345 | No |
| 53 | Homer2 | 9811 | -0.069 | -0.1351 | No |
| 54 | Camkk2 | 10330 | -0.093 | -0.1614 | No |
| 55 | Rab4a | 10985 | -0.123 | -0.1943 | No |
| 56 | H1f0 | 11078 | -0.128 | -0.1931 | No |
| 57 | Selenop | 11195 | -0.135 | -0.1930 | No |
| 58 | Cdc14b | 11563 | -0.153 | -0.2070 | No |
| 59 | Idi1 | 11666 | -0.158 | -0.2048 | No |
| 60 | Iqgap2 | 12602 | -0.206 | -0.2502 | Yes |
| 61 | Ncoa4 | 12680 | -0.212 | -0.2437 | Yes |
| 62 | Arid5b | 12939 | -0.229 | -0.2472 | Yes |
| 63 | Fkbp5 | 13142 | -0.241 | -0.2467 | Yes |
| 64 | Uap1 | 13262 | -0.250 | -0.2407 | Yes |
| 65 | Slc26a2 | 13421 | -0.261 | -0.2365 | Yes |
| 66 | Sec24d | 13499 | -0.268 | -0.2270 | Yes |
| 67 | Ccnd3 | 13741 | -0.283 | -0.2266 | Yes |
| 68 | Xrcc5 | 13904 | -0.296 | -0.2208 | Yes |
| 69 | Ank | 14029 | -0.305 | -0.2122 | Yes |
| 70 | Akt1 | 14331 | -0.328 | -0.2131 | Yes |
| 71 | Pgm3 | 14494 | -0.342 | -0.2048 | Yes |
| 72 | Maf | 14672 | -0.355 | -0.1968 | Yes |
| 73 | B2m | 14684 | -0.356 | -0.1787 | Yes |
| 74 | Pdlim5 | 14915 | -0.380 | -0.1726 | Yes |
| 75 | Dbi | 15001 | -0.391 | -0.1571 | Yes |
| 76 | Mertk | 15195 | -0.409 | -0.1472 | Yes |
| 77 | Lifr | 15599 | -0.466 | -0.1469 | Yes |
| 78 | Sord | 16054 | -0.550 | -0.1453 | Yes |
| 79 | Tpd52 | 16158 | -0.574 | -0.1213 | Yes |
| 80 | Ptpn21 | 16168 | -0.578 | -0.0914 | Yes |
| 81 | Acsl3 | 16254 | -0.608 | -0.0645 | Yes |
| 82 | Ndrg1 | 16356 | -0.640 | -0.0369 | Yes |
| 83 | Plpp1 | 16684 | -1.092 | 0.0009 | Yes |